For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AREKKNSTLAKLAGLGLAGVGVAHFAKPQLFESITKPAFPRDTRKHIYTNGGIETALGLALSASKTRRIG AVGAVGYLTYLAGNAVRNR*
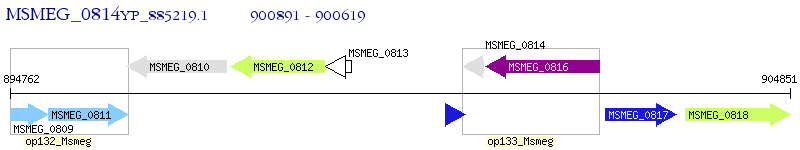
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0814 | - | - | 100% (90) | membrane protein |
| M. smegmatis MC2 155 | MSMEG_1944 | - | 1e-07 | 40.68% (59) | membrane protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0185 | - | 2e-22 | 50.59% (85) | hypothetical protein Mflv_0185 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1872c | - | 8e-18 | 50.00% (78) | hypothetical protein MAB_1872c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_2126 | - | 7e-06 | 33.78% (74) | membrane protein |
| M. thermoresistible (build 8) | TH_3144 | - | 7e-20 | 48.24% (85) | PUTATIVE - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0720 | - | 7e-26 | 58.82% (85) | hypothetical protein Mvan_0720 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0185|M.gilvum_PYR-GCK ---------------MSNE---NSRAA-------------------TVAG
Mvan_0720|M.vanbaalenii_PYR-1 ---------------MPNE---NSRAA-------------------TVAG
MSMEG_0814|M.smegmatis_MC2_155 ---------------MAREK-KNSTLA-------------------KLAG
TH_3144|M.thermoresistible__bu --------------VADRDSDKDSKVL-------------------KLSG
MAB_1872c|M.abscessus_ATCC_199 MALRRKKALKLLVDGQPTATLVTTKVGPSLFERLSVLIANLIRIGFRAGG
MAV_2126|M.avium_104 ------------MTTRSAAARDATELAAY-----------------RVAA
: ..
Mflv_0185|M.gilvum_PYR-GCK LALAATGLSHFVAPHLYEDLTKQAFPTDTRQHVYIDGAIETAVGLGIASP
Mvan_0720|M.vanbaalenii_PYR-1 LALAGAGVSHFVAPALYEGLTKPAFPTNTRQHVYVDGAIETAVGLGIASP
MSMEG_0814|M.smegmatis_MC2_155 LGLAGVGVAHFAKPQLFESITKPAFPRDTRKHIYTNGGIETALGLALSAS
TH_3144|M.thermoresistible__bu LLLAATGLSHLAAPTFWEPLVSGVFPDNTRNHVYVNGGIETALGVGLAAR
MAB_1872c|M.abscessus_ATCC_199 AGLAATGVAHFVAPQPFESISKVAFPEDTRRWVYQNGFTELLLGLALAFR
MAV_2126|M.avium_104 MLLG-VGTLHFLAPKPFDSIIPAELPGSPRLYTYGSGVAELTVGALLVPR
*. .* *: * :: : :* ..* * .* * :* :
Mflv_0185|M.gilvum_PYR-GCK KTRKFALIG-LAVYLIYMLGNVARNR------------------------
Mvan_0720|M.vanbaalenii_PYR-1 KTRRLALVG-LVVYLLYLAGNVARNR------------------------
MSMEG_0814|M.smegmatis_MC2_155 KTRRIGAVG-AVGYLTYLAGNAVRNR------------------------
TH_3144|M.thermoresistible__bu RTRKFAVVG-LLAYTAYLVATAVRSRST----------------------
MAB_1872c|M.abscessus_ATCC_199 RTRIVGSLG-GLAYVAFLVSRLVGNASKS---------------------
MAV_2126|M.avium_104 RTRRAAALAAAILFVGVFPGNLNMVRLWWDKPWPMRIAALARLPLQIPMI
:** . :. : : .
Mflv_0185|M.gilvum_PYR-GCK ----------
Mvan_0720|M.vanbaalenii_PYR-1 ----------
MSMEG_0814|M.smegmatis_MC2_155 ----------
TH_3144|M.thermoresistible__bu ----------
MAB_1872c|M.abscessus_ATCC_199 ----------
MAV_2126|M.avium_104 TTALRIRRNS