For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARVTEEQVEAVRALVAAIPPGTVSTYGDIAEVAGLSSPRIVGWIMRTDSSDLPWHRVVPASGRPAPHLAT RQLERLRAEGVLATDGRVPLREYRHIF*
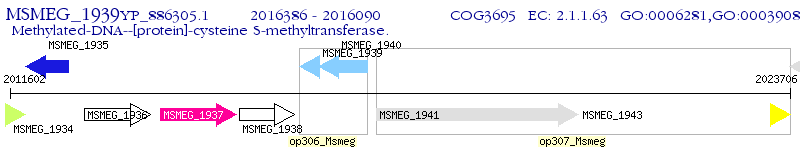
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1939 | - | - | 100% (98) | 6-O-methylguanine DNA methyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3229 | - | 8e-40 | 80.61% (98) | DNA-methyltransferase (modification methylase) |
| M. gilvum PYR-GCK | Mflv_4668 | - | 1e-42 | 80.61% (98) | methylated-DNA-(protein)-cysteine S-methyltransferase |
| M. tuberculosis H37Rv | Rv3204 | - | 2e-39 | 79.59% (98) | DNA-methyltransferase (modification methylase) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3518 | - | 1e-39 | 76.53% (98) | DNA-methyltransferase |
| M. marinum M | MMAR_1354 | - | 1e-36 | 73.47% (98) | DNA-methyltransferase (modification methylase) |
| M. avium 104 | MAV_4152 | - | 4e-41 | 81.63% (98) | 6-O-methylguanine DNA methyltransferase |
| M. thermoresistible (build 8) | TH_0448 | - | 5e-40 | 81.25% (96) | POSSIBLE DNA-METHYLTRANSFERASE (MODIFICATION METHYLASE) |
| M. ulcerans Agy99 | MUL_2525 | - | 2e-36 | 72.45% (98) | DNA-methyltransferase (modification methylase) |
| M. vanbaalenii PYR-1 | Mvan_1799 | - | 2e-45 | 85.71% (98) | methylated-DNA-(protein)-cysteine S-methyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3229|M.bovis_AF2122/97 MAPVTDEQVELVRSLVAAIPLGRVSTYGDIAALAGLSSPRIVGWIMRTDS
Rv3204|M.tuberculosis_H37Rv MAPVTDEQVELVRSLVAAIPLGRVSTYGDIAALTGLSSPRIVGWIMRTDS
MMAR_1354|M.marinum_M MAPVTDEQVERVRRLVAAIPSGRVATYGDIASAAGLSSPRIVGWIMRTDS
MUL_2525|M.ulcerans_Agy99 MAPVTDEQVERVRRLVAAIPSGRVATYGDIASAAGLSSPRIAGWIMRTDS
MAV_4152|M.avium_104 MAPVTDEQVERVRALVAAIPPGRVATYGDIAAVAGLSSARIVGWIMRTDS
MAB_3518|M.abscessus_ATCC_1997 MAAVTEEQVELVRSLVASIPAGRVATYGDIADAAGLASARIVGWIMRTDS
MSMEG_1939|M.smegmatis_MC2_155 MARVTEEQVEAVRALVAAIPPGTVSTYGDIAEVAGLSSPRIVGWIMRTDS
Mvan_1799|M.vanbaalenii_PYR-1 MAAVTEEQVETVRGLVASVPPGRVTTYGDIADAAGLSSPRIVGWIMRTDS
Mflv_4668|M.gilvum_PYR-GCK MAAITDDQVEAVRALVAAIPAGRVATYGDIAGAAGLSSPRIVAWIMRTDS
TH_0448|M.thermoresistible__bu MAPVTDEQVEAVRALVAAIPSGRVVTYGDIAAAAGLSSARTVGWIMRTDS
** :*::*** ** ***::* * * ****** :**:*.* ..*******
Mb3229|M.bovis_AF2122/97 SDLPWHRVIRASGRPAQHLATRQLELLRAEGVLSVDGRVALSEIRYEFPP
Rv3204|M.tuberculosis_H37Rv SDLPWHRVIRASGRPAQHLATRQLELLRAEGVLSVDGRVALSEIRYEFPP
MMAR_1354|M.marinum_M VDLPWHRVITASGRPARHLTNRQLELLRAEGVLSVDGKIALDQVRYEFPP
MUL_2525|M.ulcerans_Agy99 VDLPWHRVITASGRPARHLTSRQLELLRAEGVLSVDGKIALDQVRYEFPP
MAV_4152|M.avium_104 SDLPWHRVITASGRPARHLRTRQLELLRAEGVLATEGRIPLPEVRHRFGA
MAB_3518|M.abscessus_ATCC_1997 ADLPWHRVIGASGRPAPHIRTRQLELLRAEGVLAVDGRIALRDVRHGF--
MSMEG_1939|M.smegmatis_MC2_155 SDLPWHRVVPASGRPAPHLATRQLERLRAEGVLATDGRVPLREYRHIF--
Mvan_1799|M.vanbaalenii_PYR-1 SDLPWHRVIAASGRPAVHLATRQLERLRAEGVLAVDGRVPLREYRHTF--
Mflv_4668|M.gilvum_PYR-GCK ADLPWHRVIPVSGRPAPHLAGRQLDRLRAEGVLARDGRVPLTQYRHAF--
TH_0448|M.thermoresistible__bu ADLPWYRVIAASGRPAPHLAARQLELLRTEGVLAVDGRVPLRVYRHRLEI
****:**: .***** *: ***: **:****: :*::.* *: :
Mb3229|M.bovis_AF2122/97 G-
Rv3204|M.tuberculosis_H37Rv G-
MMAR_1354|M.marinum_M GC
MUL_2525|M.ulcerans_Agy99 GC
MAV_4152|M.avium_104 --
MAB_3518|M.abscessus_ATCC_1997 --
MSMEG_1939|M.smegmatis_MC2_155 --
Mvan_1799|M.vanbaalenii_PYR-1 --
Mflv_4668|M.gilvum_PYR-GCK --
TH_0448|M.thermoresistible__bu --