For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGSDSDWTVMVDAAEALAEFEVPFEVGVVSAHRTPGRMLDYAKNAADRGIEVIIAGAGGAAHLPGMVASA TPLPVIGVPVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRILGATDPQLRGRMAKFQADL EAMVLEKDAALRSRLLGDG
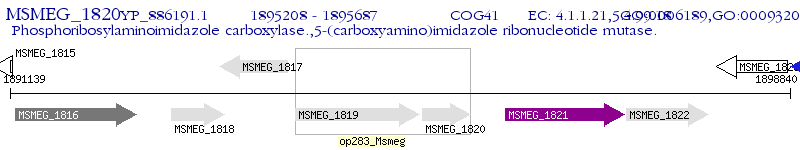
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1820 | purE | - | 100% (159) | phosphoribosylaminoimidazole carboxylase, catalytic subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3303c | purE | 2e-62 | 77.71% (157) | phosphoribosylaminoimidazole carboxylase catalytic subunit |
| M. gilvum PYR-GCK | Mflv_4746 | - | 2e-70 | 84.81% (158) | phosphoribosylaminoimidazole carboxylase, catalytic subunit |
| M. tuberculosis H37Rv | Rv3275c | purE | 1e-61 | 77.07% (157) | phosphoribosylaminoimidazole carboxylase catalytic subunit |
| M. leprae Br4923 | MLBr_00736 | purE | 3e-64 | 77.64% (161) | phosphoribosylaminoimidazole carboxylase |
| M. abscessus ATCC 19977 | MAB_3619c | - | 1e-70 | 84.81% (158) | phosphoribosylaminoimidazole carboxylase PurE |
| M. marinum M | MMAR_1261 | purE | 3e-63 | 80.26% (152) | phosphoribosylaminoimidazole carboxylase catalytic subunit |
| M. avium 104 | MAV_4244 | purE | 7e-65 | 82.24% (152) | phosphoribosylaminoimidazole carboxylase, catalytic subunit |
| M. thermoresistible (build 8) | TH_0890 | purE | 2e-73 | 89.17% (157) | PROBABLE PHOSPHORIBOSYLAMINOIMIDAZOLE CARBOXYLASE CATALYTIC |
| M. ulcerans Agy99 | MUL_2627 | purE | 2e-63 | 80.26% (152) | phosphoribosylaminoimidazole carboxylase catalytic subunit |
| M. vanbaalenii PYR-1 | Mvan_1714 | - | 2e-71 | 86.08% (158) | phosphoribosylaminoimidazole carboxylase, catalytic subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4746|M.gilvum_PYR-GCK -------------MSSPRVGLIMGSDSDWPVMSDAAEALAEFEVPFEVGV
Mvan_1714|M.vanbaalenii_PYR-1 --------------MTPRVGLIMGSDSDWPVMSEAAEALAEFDVPFEVGV
TH_0890|M.thermoresistible__bu MGMPTEERLREEQRMRPRVGLIMGSDSDWPVMSEAAEALAEFDVPFEVGV
MSMEG_1820|M.smegmatis_MC2_155 ----------------------MGSDSDWTVMVDAAEALAEFEVPFEVGV
MAB_3619c|M.abscessus_ATCC_199 --------------MSARVGLIMGSDSDWSVMSDAANALAEFEIPFEVGV
Mb3303c|M.bovis_AF2122/97 ---------MTPAGERPRVGVIMGSDSDWPVMADAAAALAEFDIPAEVRV
Rv3275c|M.tuberculosis_H37Rv ---------MTPAGERPRVGVIMGSDSDWPVMADAAAALAEFDIPAEVRV
MMAR_1261|M.marinum_M ------------MAEQPRVAVIMGSDSDWSVMADAAKALAEFDIAMEVRV
MUL_2627|M.ulcerans_Agy99 ------------MAEQPRVAVIMGSDSDWSVMADAAKALAEFDIAMEVRV
MLBr_00736|M.leprae_Br4923 ------------MTRQPRVGVIMGSDSDWSVMQDAAHALAEFDIPIEVRV
MAV_4244|M.avium_104 ----------MSVTEAPRVAVIMGSDSDWSVMADAAAALAEFDVPTEVRV
*******.** :** *****::. ** *
Mflv_4746|M.gilvum_PYR-GCK VSAHRTPARMLTYAQEASGRGIEVIIAGAGGAAHLPGMVASATPLPVIGV
Mvan_1714|M.vanbaalenii_PYR-1 VSAHRTPARMLSYAADAAGRGLEVIIAGAGGAAHLPGMVASATPLPVIGV
TH_0890|M.thermoresistible__bu VSAHRTPGRMLDYARGAAGRGIEVIIAGAGGAAHLPGMVASATPLPVIGV
MSMEG_1820|M.smegmatis_MC2_155 VSAHRTPGRMLDYAKNAADRGIEVIIAGAGGAAHLPGMVASATPLPVIGV
MAB_3619c|M.abscessus_ATCC_199 VSAHRTPGRMLQYAQSAAGRGIEVIIAGAGGAAHLPGMVASATPLPVIGV
Mb3303c|M.bovis_AF2122/97 VSAHRTPEAMFSYARGAAERGLEVIIAGAGGAAHLPGMVAAATPLPVIGV
Rv3275c|M.tuberculosis_H37Rv VSAHRTPEAMFSYARGAAERGLEVIIAGAGGAAHLPGMVAAATPLPVIGV
MMAR_1261|M.marinum_M VSAHRTPQVMFDYARDAAGRGIEVIIAGAGGAAHLPGMVASATPLPVIGV
MUL_2627|M.ulcerans_Agy99 VSAHRTPQVMFDYARDAAGRGIEVIIAGAGGAAHLPGMVASATPLPVIGV
MLBr_00736|M.leprae_Br4923 VSAHRTPAEMFDYARNAVDRSIAVIIAGAGGAAHLPGMVASATPLPVIGV
MAV_4244|M.avium_104 VSAHRTPGVMFDYARDAAGRGIEVIIAGAGGAAHLPGMVASATPLPVIGV
******* *: ** * *.: *****************:*********
Mflv_4746|M.gilvum_PYR-GCK PVPLARLDGMDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRILGASDNA
Mvan_1714|M.vanbaalenii_PYR-1 PVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRILGAADGA
TH_0890|M.thermoresistible__bu PVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRILAAADSA
MSMEG_1820|M.smegmatis_MC2_155 PVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRILGATDPQ
MAB_3619c|M.abscessus_ATCC_199 PVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAARILGVADTA
Mb3303c|M.bovis_AF2122/97 PVPLGRLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRMLGAANPQ
Rv3275c|M.tuberculosis_H37Rv PVPLGRLDGLDSLLSIVQMPAGVPVATVSIGGAGNAGLLAVRMLGAANPQ
MMAR_1261|M.marinum_M PVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRMLGSSDPA
MUL_2627|M.ulcerans_Agy99 PVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRMLGSSDPA
MLBr_00736|M.leprae_Br4923 PVPLARLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRILGSSDLQ
MAV_4244|M.avium_104 PVPLGRLDGLDSLLSIVQMPAGVPVATVSIGGARNAGLLAVRILGASDPG
****.****:*********************** ******.*:*. ::
Mflv_4746|M.gilvum_PYR-GCK LQQRMIAFQASLEQMVLQKEEALR---QKLLGE----
Mvan_1714|M.vanbaalenii_PYR-1 LRDRMAAYQASLEQMVLQKDEALR---QKLLGD----
TH_0890|M.thermoresistible__bu LRDRMTKFQADLERMVLAKDEALR---DRLLGGDRKE
MSMEG_1820|M.smegmatis_MC2_155 LRGRMAKFQADLEAMVLEKDAALR---SRLLGDG---
MAB_3619c|M.abscessus_ATCC_199 LRDKVVAFQASMEATVLEKDEALR---RRLLED----
Mb3303c|M.bovis_AF2122/97 LRARIVAFQDRLADVVAAKDAELQRLAGKLTRD----
Rv3275c|M.tuberculosis_H37Rv LRARIVAFQDRLADVVAAKDAELQRLAGKLTRD----
MMAR_1261|M.marinum_M LRARIAAFQDQLAQTVQAKDAELQKLAGKLTRD----
MUL_2627|M.ulcerans_Agy99 LRARIAAFQDQLAQTVQAKDAELQKLAGKLTRD----
MLBr_00736|M.leprae_Br4923 LRAQLVAFQDRLADTVRAKDADLQRFRGKLIGD----
MAV_4244|M.avium_104 LRARIVEFQTQLADSVRAKDEALQRLHGKVTGQ----
*: :: :* : * *: *: ::