For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LSLGGNPKWMGLPSSQRPPLTMVHPERLPGAPGPYTAGRRRVVVEVPAREAMLAPLRNILGAIATFEGLD SDVVADLCLAVDEACAVLINAAAPDSLVSLDIVADAQMLVVDASTTCSRPEDAGLGRFSERVLSALTDEV GTFAAAADTANGTEDDPGNSPGNDPENGTDPRGVFGISLTTRRRPPTASDAVSD
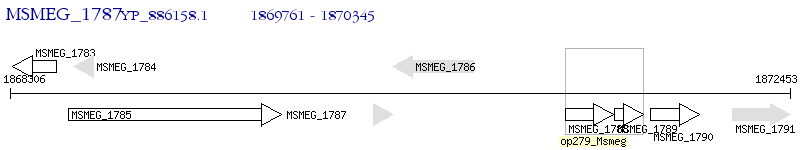
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1787 | - | - | 100% (194) | RsbW protein |
| M. smegmatis MC2 155 | MSMEG_1803 | - | 2e-22 | 43.84% (146) | RsbW protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3315c | rsbW | 7e-20 | 42.47% (146) | anti-sigma factor rsbW (sigma negative effector) |
| M. gilvum PYR-GCK | Mflv_4790 | - | 3e-21 | 45.77% (142) | putative anti-sigma regulatory factor, serine/threonine protein |
| M. tuberculosis H37Rv | Rv3287c | rsbW | 7e-20 | 42.47% (146) | anti-sigma factor rsbW (sigma negative effector) |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2513c | - | 3e-12 | 36.45% (107) | anti-sigma factor RsbW |
| M. marinum M | MMAR_1247 | rsbW | 2e-19 | 41.78% (146) | anti-sigma factor RsbW |
| M. avium 104 | MAV_4258 | - | 3e-20 | 40.41% (146) | RsbW protein |
| M. thermoresistible (build 8) | TH_0240 | rsbW | 1e-15 | 39.06% (128) | ANTI-SIGMA FACTOR RSBW (SIGMA NEGATIVE EFFECTOR) |
| M. ulcerans Agy99 | MUL_2641 | rsbW | 8e-21 | 43.15% (146) | anti-sigma factor RsbW |
| M. vanbaalenii PYR-1 | Mvan_1659 | - | 1e-21 | 43.15% (146) | putative anti-sigma regulatory factor, serine/threonine protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1247|M.marinum_M MTNRLQRWAQSGTQSESTVGAKLPGGQMTDADPRINKR---ERGHRAVEL
MUL_2641|M.ulcerans_Agy99 ---------------------------MTDADPRINKR---ERGHRAVEL
MAV_4258|M.avium_104 ---------------------------MKDAGLHADKR---PRGHRAVEL
Mb3315c|M.bovis_AF2122/97 ---------------------------MADSDLPTKGR---QRGVRAVEL
Rv3287c|M.tuberculosis_H37Rv ---------------------------MADSDLPTKGR---QRGVRAVEL
Mflv_4790|M.gilvum_PYR-GCK --------MDSGDKAWGNLASKFPGGQMADVASHSNGQ---RLSPQSVEL
Mvan_1659|M.vanbaalenii_PYR-1 --------MDNGRSGWGNLAGKLLGGQMADVASHINGR---RQGAQSVEL
TH_0240|M.thermoresistible__bu --------------------------------------------------
MAB_2513c|M.abscessus_ATCC_199 ---------------------------MTEAHTTHSGS---HPDDRSLRF
MSMEG_1787|M.smegmatis_MC2_155 ------MSLGGNPKWMGLPSSQRPPLTMVHPERLPGAPGPYTAGRRRVVV
MMAR_1247|M.marinum_M NVAARLENLSMLRTLVGAIGTFEDLDFDAVADLRLAVDEVCTRLIRSATP
MUL_2641|M.ulcerans_Agy99 NVAARLENLSMLRTLVGAIGTFEDLDFDAVADLRLAVDEVCTRLIRSATP
MAV_4258|M.avium_104 HVAARLENLAMLRTLVGAIGTFEDLDFDAVADLRLAVDEVCTRLIRSATP
Mb3315c|M.bovis_AF2122/97 NVAARLENLALLRTLVGAIGTFEDLDFDAVADLRLAVDEVCTRLIRSALP
Rv3287c|M.tuberculosis_H37Rv NVAARLENLALLRTLVGAIGTFEDLDFDAVADLRLAVDEVCTRLIRSALP
Mflv_4790|M.gilvum_PYR-GCK RVAAALENLAVLRTLVAAIGTFEDLDFDAVADLRLAVDEACTRLIRSALP
Mvan_1659|M.vanbaalenii_PYR-1 RVAATLENLAVLRTLVAAIGTFEDLDFDAVADLRLAVDEACTRLIRSAVP
TH_0240|M.thermoresistible__bu ----------VLRTLVAAVGTFEDLDFDAVADLRLAVDEACTRLVRSAVP
MAB_2513c|M.abscessus_ATCC_199 DMAARLENLAMMRTVVAAAATVDDLDMDMVADLKLATDEACTRLVRSSVP
MSMEG_1787|M.smegmatis_MC2_155 EVPAREAMLAPLRNILGAIATFEGLDSDVVADLCLAVDEACAVLINAAAP
:*.::.* .*.:.** * **** **.**.*: *:.:: *
MMAR_1247|M.marinum_M DATLTLVVDPRDDELVVQASAACGTHDVVAPGSFSWHVLTSLADDVQTFH
MUL_2641|M.ulcerans_Agy99 DATLTLVVDPRDDELVVQASAACSTHDVVALGSFSWHVLTSLADDVQTFH
MAV_4258|M.avium_104 DATLIVVVDPQDDQLVVEASAACDTHDVVAPGSFSWHVLTSLADDVQTFH
Mb3315c|M.bovis_AF2122/97 DATLRLVVDPRKDEVVVEASAACDTHDVVAPGSFSWHVLTALADDVQTFH
Rv3287c|M.tuberculosis_H37Rv DATLRLVVDPRKDEVVVEASAACDTHDVVAPGSFSWHVLTALADDVQTFH
Mflv_4790|M.gilvum_PYR-GCK DSTLLVVVDPQPDALVVHASTTCDSTDILAPGSFSWHVLSSLTDEVTTFS
Mvan_1659|M.vanbaalenii_PYR-1 DSTLLVVVDPQPDALVVHASTTCDSTDILAPGSFSWHVLSSLTDDVKTFS
TH_0240|M.thermoresistible__bu GATLVVRVHPYDDVVVVEASTTVTETDILSPGSFSWHVLNSLTDEVRTFQ
MAB_2513c|M.abscessus_ATCC_199 NAMLVVRLDFHLDRLVMAVYSHCQPGDVFPLDSFSWHVLSSLADHVETFE
MSMEG_1787|M.smegmatis_MC2_155 DSLVSLDIVADAQMLVVDASTTCSRPEDAGLGRFSERVLSALTDEVGTFA
.: : : : : :*: . : : . ** :**.:*:*.* **
MMAR_1247|M.marinum_M ------DGRPPEA-------------AGSVFGIMLTARRVASGR------
MUL_2641|M.ulcerans_Agy99 ------DGRPPEA-------------AGSVFGIMLTARRVASGR------
MAV_4258|M.avium_104 ------DGREPNE-------------AGSVFGITLTARRAASSR------
Mb3315c|M.bovis_AF2122/97 ------DGRQPDV-------------AGSVFGITLTARRAASSR------
Rv3287c|M.tuberculosis_H37Rv ------DGRQPDV-------------AGSVFGITLTARRAASSR------
Mflv_4790|M.gilvum_PYR-GCK ------DGQAAD--------------DGEVFGISMTTRRASSLQ------
Mvan_1659|M.vanbaalenii_PYR-1 ------DGRSVG--------------DGEVFGISMTTRRASSLQ------
TH_0240|M.thermoresistible__bu ------DGQQAS--------------GAQVCGIAMTTRRASSLR------
MAB_2513c|M.abscessus_ATCC_199 ------RAHPDDP-------------VGHIVGISLTKLRDTGSAVRVANG
MSMEG_1787|M.smegmatis_MC2_155 AAADTANGTEDDPGNSPGNDPENGTDPRGVFGISLTTRRRPPTASDAVSD
. : ** :* * .