For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LRRPAASPKGLPERPGHEGIGPARLKVQGGRLVDELERRFGAGWKVRAGEVFSRDGTVLDESTVLPPGSH VYLYRDLPDEVDVPFDIPVLYRDDDIVVVDKPHFLATMPRGGHVTQTALVRLRRELDLPELAPAHRLDRL TAGVLLFTVRRELRGAYQTMFARGDVQKTYLAHAEGVPKIALPATLRSRIVKHRGRLQAFEEPGEPNAET YVEAVGDGLYRLTPRTGRTHQLRVHMASIGLPITGDPFYPTVIDVAPGDFSRPLQLLAKRLMFDDPVSGA RREFVSRRTLG
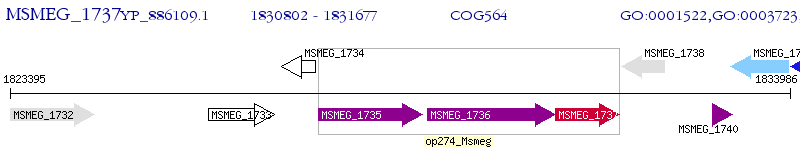
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1737 | - | - | 100% (291) | RNA pseudouridine synthase family protein |
| M. smegmatis MC2 155 | MSMEG_3175 | - | 3e-11 | 29.09% (220) | ribosomal large subunit pseudouridine synthase D |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3328c | - | 1e-107 | 70.76% (277) | hypothetical protein Mb3328c |
| M. gilvum PYR-GCK | Mflv_4820 | - | 1e-118 | 71.38% (290) | pseudouridine synthase |
| M. tuberculosis H37Rv | Rv3300c | - | 1e-107 | 70.76% (277) | hypothetical protein Rv3300c |
| M. leprae Br4923 | MLBr_01200 | - | 1e-14 | 31.46% (213) | putative pseudouridine synthase |
| M. abscessus ATCC 19977 | MAB_3654c | - | 7e-82 | 60.22% (269) | pseudouridine synthase |
| M. marinum M | MMAR_1231 | - | 1e-102 | 67.38% (279) | pseudouridine synthase |
| M. avium 104 | MAV_4277 | - | 1e-106 | 69.89% (279) | RNA pseudouridine synthase family protein |
| M. thermoresistible (build 8) | TH_1839 | - | 6e-95 | 67.43% (261) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_2659 | - | 1e-101 | 67.38% (279) | pseudouridine synthase |
| M. vanbaalenii PYR-1 | Mvan_1621 | - | 1e-106 | 67.47% (292) | pseudouridine synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3328c|M.bovis_AF2122/97 MALRPEDRL--LSVH---DVLGPVRVRLL-GGSVLAELTARFGVAARAKV
Rv3300c|M.tuberculosis_H37Rv MALRPEDRL--LSVH---DVLGPVRVRLL-GGSVLAELTARFGVAARAKV
MMAR_1231|M.marinum_M --MKPAP----LRVR---DALGPARVRLH-GGPVLAELSERFGAAARAKV
MUL_2659|M.ulcerans_Agy99 --MKPAP----LRVR---DALGPARVRLH-GGPVLAELSERFGAAARAKV
MAV_4277|M.avium_104 -----MP----LPVR---DGLGPARVRLR-GGPVLAELTARFGAAARAKV
MSMEG_1737|M.smegmatis_MC2_155 -MRRPAASPKGLPERPGHEGIGPARLKVQ-GGRLVDELERRFG--AGWKV
Mflv_4820|M.gilvum_PYR-GCK -MRRPPP----LPVR---DGLGPARVRVQ-GGLLVEEFQRRWG--TGAKV
Mvan_1621|M.vanbaalenii_PYR-1 -MRRPAP----LPVR---DGLGPARVRLR-GGAVLVELADRFGEAAAAKV
TH_1839|M.thermoresistible__bu ---------------------------------VLVEFAERFGEAAAAKV
MAB_3654c|M.abscessus_ATCC_199 -----------------------MRIRLHDGGRVVDELVARFGPAAVDQV
MLBr_01200|M.leprae_Br4923 MTDRSMPIP---------EGLAGMRVDAGLARLLGLSRTAVAALAEDGGV
: . . *
Mb3328c|M.bovis_AF2122/97 LAGEVVDNDGAVVDSGTVLPPGSVVHLYRDLPDEVPVPFDVPVLHQDADI
Rv3300c|M.tuberculosis_H37Rv LAGEVVDDDGAVVDSGTVLPPGSVVHLYRDLPDEVPVPFDVPVLHQDADI
MMAR_1231|M.marinum_M LAGEVVDAAGAIVDTATVAEPGSYVYLYRDLPEEVPVPFEMPVLHQDEDI
MUL_2659|M.ulcerans_Agy99 LAGEVVDAAGAIVGTATVAGPGSHVYLYRDLPEEVPVPFEMPVLHQDEDI
MAV_4277|M.avium_104 LAAEVVTADGAVVDEATVLPAGASVYLYRELPDEVVVPFDLPVLHRDDDI
MSMEG_1737|M.smegmatis_MC2_155 RAGEVFSRDGTVLDESTVLPPGSHVYLYRDLPDEVDVPFDIPVLYRDDDI
Mflv_4820|M.gilvum_PYR-GCK VDGEVFCADGTVADGSTVLPPGAYVYLYRDLPDEVPVPFDIPVLHRDDDI
Mvan_1621|M.vanbaalenii_PYR-1 LNGEVFTADGAVVTASTVLQPNSVVYLYRDLPDEVAVPFDVPVLHRDDNL
TH_1839|M.thermoresistible__bu LRGEVFCADGTPVTADTVLPPGSVVYLYRELPEEVPVPFDIPILYRDENI
MAB_3654c|M.abscessus_ATCC_199 DRGEVVDSAGMRVTAGTRLPAGAVVYTYREPAEEISVPFAIDVLHWDENV
MLBr_01200|M.leprae_Br4923 ELDGVQAGKSDRLTSGAWLQVR-LCEAPAPLENTPVDIEDMTILYSDDDI
* . : : : :*: * ::
Mb3328c|M.bovis_AF2122/97 VVVDKPHFLATMPRGRHVAQTALVRLR------RELGLPELS-PAHRLDR
Rv3300c|M.tuberculosis_H37Rv VVVDKPHFLATMPRGRHVAQTALVRLR------RELGLPELS-PAHRLDR
MMAR_1231|M.marinum_M VVADKPHFLATMPRGRHVAQTALVRLR------CELGLPELS-PAHRLDR
MUL_2659|M.ulcerans_Agy99 VVADKPHFLATMPRGRHVAQTALVRLR------CELGLPELS-PAHRLDR
MAV_4277|M.avium_104 VVVDKPHFLATMPRGRHVAQTALVRLR------RELGLPELS-PAHRLDR
MSMEG_1737|M.smegmatis_MC2_155 VVVDKPHFLATMPRGGHVTQTALVRLR------RELDLPELA-PAHRLDR
Mflv_4820|M.gilvum_PYR-GCK VVVDKPHFLATMPRGSHVAQTALVRLR------RELDLPELS-PAHRLDR
Mvan_1621|M.vanbaalenii_PYR-1 VVVDKPHFLATMPRGSHVAQTALVRMR------RELDLPQLS-PAHRLDR
TH_1839|M.thermoresistible__bu VVVDKPHFLATTPRGRHVAQTALVRLR------RQLDLPELS-PAHRLDR
MAB_3654c|M.abscessus_ATCC_199 LVVDKPHFLATMPRGRHVTETALVRLR------KMLDLPELA-PAHRLDR
MLBr_01200|M.leprae_Br4923 VAVDKPAAVAAHASVGWAGPTVLGGLAAAGYRITTSGVPERQGIVHRLDV
:..*** :*: . . *.* : .:*: .****
Mb3328c|M.bovis_AF2122/97 LTAGVLLFTTRREVRGSYQTMFARGLVRKTYLARAPVAPGLALPRLVRSR
Rv3300c|M.tuberculosis_H37Rv LTAGVLLFTTRREVRGSYQTMFARGLVRKTYLARAPVAPGLALPRLVRSR
MMAR_1231|M.marinum_M LTAGVLLFTTRRELRGRYQTLFARGAVRKTYLARAAVDADLAFPRVIRSR
MUL_2659|M.ulcerans_Agy99 LTAGVLLFTTRRELRGRYQTLFARGAVRKTYLARAAVDADLAFPRVIRSR
MAV_4277|M.avium_104 LTAGVLLFTARREVRGAYQTLFSRGLVDKTYLARAAVDPALVFPRVLAGR
MSMEG_1737|M.smegmatis_MC2_155 LTAGVLLFTVRRELRGAYQTMFARGDVQKTYLAHAEGVPKIALPATLRSR
Mflv_4820|M.gilvum_PYR-GCK LTAGVLLFTVRREVRGAYQTLFVRGEAAKTYVAHSSHATDREFPMTIRNR
Mvan_1621|M.vanbaalenii_PYR-1 LTAGVLVFTTRREVRGAYQTMFARGGVSKTYLAQSSAGTDRDFPITVRSR
TH_1839|M.thermoresistible__bu LTAGVLLFTTRREVRGRYQRLFADEAVRKIYLAGAPVRPDLEFPRVVRSR
MAB_3654c|M.abscessus_ATCC_199 LTAGVLLFTVRRDARAAYHSLFAQRQVVKIYEALAPVKPGLSLPLTVRSR
MLBr_01200|M.leprae_Br4923 GTSGVMLVAISERAYTVLKRAFKCRTVDKRYHALVQGHPDPSS-GTIDAP
*:**::.: . : * . * * * . :
Mb3328c|M.bovis_AF2122/97 IVKRRGHLQAVCEPGV-PNAETLVERIAR---DGLYRLTPTTGRTHQLRV
Rv3300c|M.tuberculosis_H37Rv IVKRRGHLQAVCEPGV-PNAETLVERIAR---DGLYRLTPTTGRTHQLRV
MMAR_1231|M.marinum_M IVKRRGHLQAVCEPGE-PNAETLVELISP---EGLYRLTPRTGRTHQLRV
MUL_2659|M.ulcerans_Agy99 IVKRRGHLQAVCEPGE-PNAETLVELISP---EGLYRLTPRTGRTHQLRV
MAV_4277|M.avium_104 IVKHRGRMQAVCEPGP-PNALTEVELLSP---DGLYRLRPRTGRTHQLRL
MSMEG_1737|M.smegmatis_MC2_155 IVKHRGRLQAFEEPGE-PNAETYVEAVG----DGLYRLTPRTGRTHQLRV
Mflv_4820|M.gilvum_PYR-GCK LVKHRGSLQAVEEPGE-PNAETVVEYLD----EGMYRLTPRTGRTHQLRV
Mvan_1621|M.vanbaalenii_PYR-1 IIKRRGTLQAVEEPGE-PNAETLVERVD----DRTYRLMPRTGRTHQLRV
TH_1839|M.thermoresistible__bu IIKRRGSLQAVEEPGE-PNAETLVELLDAS--AGRYRLTPRTGRTHQLRV
MAB_3654c|M.abscessus_ATCC_199 IVKRRSILQAVEEPGP-ANAETRVELVDRHHAVGRYRLTPSTGQTHQLRV
MLBr_01200|M.leprae_Br4923 IGRHRGHTWKFAVTKDGRHSITHYDTLEAFVAASLLDVHLETGRTHQIRA
: ::*. . . :: * : : : **:***:*
Mb3328c|M.bovis_AF2122/97 HMAALGIPIMGDPLYPNVISVAAH---DFSTPLQLLAQRIEFDDPLTGSH
Rv3300c|M.tuberculosis_H37Rv HMAALGIPIMGDPLYPNVISVAAH---DFSTPLQLLAQRIEFDDPLTGSH
MMAR_1231|M.marinum_M HMASLGLPIFGDPLYPNVIPESPHDLSDFSSPLSLLAQRIEFEDPLSGSR
MUL_2659|M.ulcerans_Agy99 HMASLGLPIFGDPLYPNVIPESPHDLSDFSSPLSLLAQRIEFEDPLSGSR
MAV_4277|M.avium_104 QMASLGIPIIGDPLYPNMIDVPAD---DFSTPLQLLAQRIEFDDPRTGAR
MSMEG_1737|M.smegmatis_MC2_155 HMASIGLPITGDPFYPTVIDVAPG---DFSRPLQLLAKRLMFDDPVSGAR
Mflv_4820|M.gilvum_PYR-GCK HMAGLGIPITGDPLYPDVVDVPPG---DFGSPLRLLAQALEFTDPISGEL
Mvan_1621|M.vanbaalenii_PYR-1 HMNSLGLPIAGDPLYPNVIDVAVG---DFSTPLKLLAQCLEFTDPLAGVR
TH_1839|M.thermoresistible__bu HMAVLGVPIVNDPLYPEVVDVAPD---DFSSPLQLLAHSVEFTDPLTGRT
MAB_3654c|M.abscessus_ATCC_199 HMNSMGVPILNDPLYPDVVEVPPD---DFTRPLQLLARTLAFRDPLNGKD
MLBr_01200|M.leprae_Br4923 HFAELHRPCCGDLVYGADPKLAKRLG---LERQWLHARSLSFAHPADGRR
:: : * .* .* . * *: : * .* *
Mb3328c|M.bovis_AF2122/97 REFASTRTLTGATLPTWSAAADCRP
Rv3300c|M.tuberculosis_H37Rv REFASTRTLTGATLPTWSAAADCRP
MMAR_1231|M.marinum_M REFVSPREI----------------
MUL_2659|M.ulcerans_Agy99 REFVSPREI----------------
MAV_4277|M.avium_104 RVFVSRRTLDDPGP-----------
MSMEG_1737|M.smegmatis_MC2_155 REFVSRRTLG---------------
Mflv_4820|M.gilvum_PYR-GCK RRFTSGRTLAGE-------------
Mvan_1621|M.vanbaalenii_PYR-1 LRFTSERTL----------------
TH_1839|M.thermoresistible__bu HRFVSARSIGGQ-------------
MAB_3654c|M.abscessus_ATCC_199 VRYESGRALGQ--------------
MLBr_01200|M.leprae_Br4923 VEIVSSYPPDLQHALDVLRAES---
*