For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRKLTWLAALLAALAMAMTLSGCGRSAEGGGGGDGDAKGTVGIAMPTKSSERWVADGQNMVDQFKALGYD TDLQYGDDVVQNQVSQIENMITKGVKLLVIAPIDGSSLTNTLQHAADLKIPVISYDRLIKGTPNVDYYAT FDNTKVGVLQANYIVDTLGVADGKGPFNLELFAGSPDDNNATYFFQGAMSVLQPYIDSGKLVVKSGQTTF DQIATLRWDGGLAQSRMDNLLSQAYTSGRVDAVLSPYDGISRGVISALKSAGYGNAAKPLPIVTGQDAEL ASVKSIVAGEQTQTVFKDTRELAKAAVQEADAVLTGGTPQVNDTETYDNGVKVVPSYLLDPVSVDKSNYK KVLIDSGYYTETQVQ
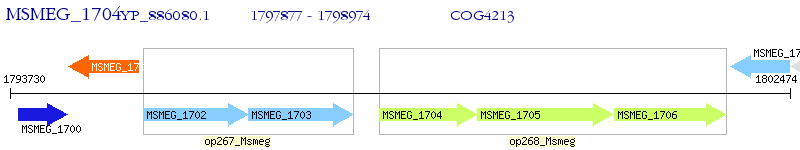
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1704 | - | - | 100% (365) | ABC transporter |
| M. smegmatis MC2 155 | MSMEG_6020 | - | 4e-45 | 35.75% (358) | D-xylose-binding periplasmic protein |
| M. smegmatis MC2 155 | MSMEG_3598 | - | 1e-14 | 25.07% (339) | periplasmic sugar-binding proteins |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00398 | - | 3e-08 | 26.16% (237) | putative D-ribose-binding protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5293 | - | 3e-42 | 35.63% (348) | solute-binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1704|M.smegmatis_MC2_155 MRKLTWLAALLAALAMAMTLSGCGRSAEGGGGGDGDAKGTVGIAMP-TKS
Mvan_5293|M.vanbaalenii_PYR-1 -MKRTSSLLFTAVVGVGLTLTACGSNSDSGS--TEARSGKVGVILPDTKS
MLBr_00398|M.leprae_Br4923 --MSTGTSVPLRVASIMLILGLLPAILPACGFSESHHKLTIGVSYP-TAN
* . .: : * . . . .:*: * * .
MSMEG_1704|M.smegmatis_MC2_155 SERWVADGQ-NMVDQFKALGYDTDLQYGDDVVQNQVSQIENMITKGVKLL
Mvan_5293|M.vanbaalenii_PYR-1 SVRWETKDRPALAAAFQNAGVDYTIQNAEGSADTMATIADGMIADGVTVL
MLBr_00398|M.leprae_Br4923 SPFWNAYTR-FIDEGSHQLGVYINAVSAEEDEQKQLSDVETLINQGIDGL
* * : : : : * .: :. : : :* .*: *
MSMEG_1704|M.smegmatis_MC2_155 VIAPIDGSSLTNTLQHAADLKIPVISYDR---LIKGTPNVDYYATFDNTK
Mvan_5293|M.vanbaalenii_PYR-1 AIVNLDSDSGASIQQKAASQGVKTIDYDR---LTLGG-SADVYVSFDNNV
MLBr_00398|M.leprae_Br4923 IVTPQSTSIAPTLLRIATQANIPVVVVDRYPGYAPGQNKNADYVAFLGPN
:. . . .. : *:. : .: ** * . *.:* .
MSMEG_1704|M.smegmatis_MC2_155 VGVLQANYIVDTLGVADGKGPFNLELFAGSPDDNNATYFFQGAMSVLQPY
Mvan_5293|M.vanbaalenii_PYR-1 VGELQGQGLVDCLGGR----PANVVFLNGSPTDNNATLFSSGAHSVVD--
MLBr_00398|M.leprae_Br4923 N-EKAGSGIAEALMAG---GGSKFLALGGMPGNSVAQGRKAGLESALT--
.. :.: * :. : * * :. * * *.:
MSMEG_1704|M.smegmatis_MC2_155 IDSGKLVVKSGQTTFDQIATLRWDGGLAQSRMDNLLSQAYTSGRVDAVLS
Mvan_5293|M.vanbaalenii_PYR-1 -------ATPSITIVGEQAVPDWDNDKAVTIFEQLYTAAD--GRVDGVYA
MLBr_00398|M.leprae_Br4923 -------AVGHRLVQFQYAGDSEDKGLAAAEN---MLQAHPAGDANAIWC
. : * * . * : * * .:.: .
MSMEG_1704|M.smegmatis_MC2_155 PYDGISRGVISALKSAGYGNAAKPLPIVTGQDAELASVKSIVAGEQTQTV
Mvan_5293|M.vanbaalenii_PYR-1 ANDGLAGSVISILEKNKRAGQVP----VTGQDATVEGLQNILAGTQCMTV
MLBr_00398|M.leprae_Br4923 FNDKLCQGAIKAVYNANREKEFI----FGGMDLTPQAIAAIENGLYTVSL
* :. ..*. : . . * * .: * * ::
MSMEG_1704|M.smegmatis_MC2_155 FKDTRELAKAAVQEADAVLTGGTPQVNDTETYDNGVKVVPSYLLDPVSVD
Mvan_5293|M.vanbaalenii_PYR-1 YKSATEEANALAEVAIALANGEQPQTTSTSRDDTGGRDVPSVLLTPKSIT
MLBr_00398|M.leprae_Br4923 GAHWLEGG-FGLAILYRKIHGQDPAERMVKLDLLKVDKSNVAKFKARYID
* . * * .. : . :
MSMEG_1704|M.smegmatis_MC2_155 KSNYKKVLIDSGYYTETQVQ------------
Mvan_5293|M.vanbaalenii_PYR-1 KDNINVVFDDGGQSKDEVCSGQFAEACSAAGV
MLBr_00398|M.leprae_Br4923 NTPHYNWKSTTPFDMTITLN------------
: .