For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSSGSKRVVFSGAQPTSDSLHLGNALGAVTHWAQLQDDYDAFFCVVDLHAITVPQDPETLRRRTLVTAAQ YLALGIDPARSTVFVQSHVPTHSQLAWVLGCFTGFGQASRMTQFKDKSQKQGADATTVGLFTYPVLMAAD VLLYDTHVVPVGEDQRQHLELARDLAQRFNARFPDTFVVPEPMIPKATAKIYDLQDPTAKMSKSAATDAG LISLLDDPKATAKKIRSAVTDSEREIRFDPEAKPGISNLLTIQSAVTGTTVDKLVESYAGRGYGDLKKDT AEAVVEFVTPIKTRVDDLLSDRAELESVLAAGARRATEVSAKTLQRVYDRIGFLPQPS
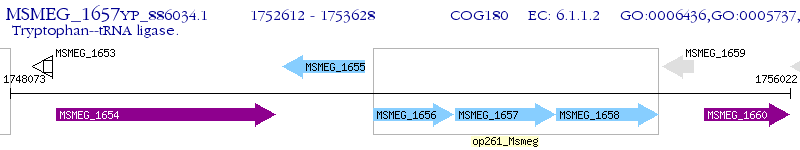
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1657 | trpS | - | 100% (338) | tryptophanyl-tRNA synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3369c | trpS | 1e-154 | 79.52% (332) | tryptophanyl-tRNA synthetase |
| M. gilvum PYR-GCK | Mflv_4871 | - | 1e-164 | 85.93% (334) | tryptophanyl-tRNA synthetase |
| M. tuberculosis H37Rv | Rv3336c | trpS | 1e-154 | 79.52% (332) | tryptophanyl-tRNA synthetase |
| M. leprae Br4923 | MLBr_00686 | trpS | 1e-152 | 78.61% (332) | tryptophanyl-tRNA synthetase |
| M. abscessus ATCC 19977 | MAB_3683c | - | 1e-138 | 70.86% (326) | tryptophanyl-tRNA synthetase |
| M. marinum M | MMAR_1188 | trpS | 1e-157 | 80.84% (334) | tryptophanyl-tRNA synthetase TrpS |
| M. avium 104 | MAV_4310 | trpS | 1e-157 | 80.24% (339) | tryptophanyl-tRNA synthetase |
| M. thermoresistible (build 8) | TH_1974 | trpS | 1e-165 | 85.33% (334) | PROBABLE TRYPTOPHANYL-TRNA SYNTHETASE TRPS (TRYPTOPHAN--TRNA |
| M. ulcerans Agy99 | MUL_1441 | trpS | 1e-158 | 81.14% (334) | tryptophanyl-tRNA synthetase |
| M. vanbaalenii PYR-1 | Mvan_1560 | - | 1e-160 | 84.73% (334) | tryptophanyl-tRNA synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4871|M.gilvum_PYR-GCK ----MSNTT-RPVVFSGAQPTSDSLHLGNALGAVSQWVSLQDGY----DA
Mvan_1560|M.vanbaalenii_PYR-1 MNTTMSNTP-RPVVFSGAQPTSDSLHLGNALGAVAQWVGLQEGN----DA
MSMEG_1657|M.smegmatis_MC2_155 ----MSSGS-KRVVFSGAQPTSDSLHLGNALGAVTHWAQLQDDY----DA
TH_1974|M.thermoresistible__bu --MNTSVRT-RPVVFSGAQPTSDSLHLGNALGAIAQWVGLQDDH----DA
MLBr_00686|M.leprae_Br4923 ----MSTATGFSRIFSGVQPTSDSLHLGNALGAITQWVALQYDHPDEYEA
MAV_4310|M.avium_104 ----MSTATGSRRIFSGVQPTSDSLHLGNALGAISQWVTLQDDWGDDWDA
Mb3369c|M.bovis_AF2122/97 ----MSTPTGSRRIFSGVQPTSDSLHLGNALGAVAQWVGLQDDH----DA
Rv3336c|M.tuberculosis_H37Rv ----MSTPTGSRRIFSGVQPTSDSLHLGNALGAVAQWVGLQDDH----DA
MMAR_1188|M.marinum_M ----MSTPTGSRRIFSGVQPTSDSLHLGNALGAVTQWVALQDDH----DA
MUL_1441|M.ulcerans_Agy99 ----MSTPTGSRRIFSGVQPTSDSLHLGNALGAVTQWVALQDDH----DA
MAB_3683c|M.abscessus_ATCC_199 ---MTAETAARGRLFSGIQPTADSLHLGNVLGAVQQWVRLQHEY----EA
: . :*** ***:*******.***: :*. ** :*
Mflv_4871|M.gilvum_PYR-GCK YFCVVDLHAITVAQDPATLRRRTLVTAAQYLALGIDPSRATVFVQSHVPS
Mvan_1560|M.vanbaalenii_PYR-1 YFCVVDLHAITVSQDPETLRKRTLATAAQYLALGVDPDRATVFVQSHVPA
MSMEG_1657|M.smegmatis_MC2_155 FFCVVDLHAITVPQDPETLRRRTLVTAAQYLALGIDPARSTVFVQSHVPT
TH_1974|M.thermoresistible__bu FFCVVDLHAITVPQDPETLRRRTLVTAAQYLALGIDPARSTVFVQSHVAA
MLBr_00686|M.leprae_Br4923 FFCVVDLHAITIAQDPETLWRRTLVTAAQYLALGIDPGRAVVFVQSHVPA
MAV_4310|M.avium_104 FFCVVDLHAITIAQDPAALRRRTLVTAAQYLALGIDPARSTVFVQSHVPA
Mb3369c|M.bovis_AF2122/97 FFCVVDLHAITIPQDPEALRRRTLITAAQYLALGIDPGRATIFVQSQVPA
Rv3336c|M.tuberculosis_H37Rv FFCVVDLHAITIPQDPEALRRRTLITAAQYLALGIDPGRATIFVQSQVPA
MMAR_1188|M.marinum_M FFCVVDLHAITLPQDPEALRRRTLVTAAQYLALGIDPARATIFVQSHVPA
MUL_1441|M.ulcerans_Agy99 FFCVVDLHAITLPQDPEALRRRTLVTAAQYLALGIDPARATIFVQSHVPA
MAB_3683c|M.abscessus_ATCC_199 LFCVVDLHAITVAQDPEELRRRTRAVVAQYVALGVDPAKSAIFVQSHVPA
**********:.*** * :** ..***:***:** ::.:****:*.:
Mflv_4871|M.gilvum_PYR-GCK HAELAWVLGCFTGFGQASRMTQFKDKSQKQGAEATTVGLFTYPVLMAADV
Mvan_1560|M.vanbaalenii_PYR-1 HAELAWVLGCFTGFGQASRMTQFKDKSQKQGADSTTVGLFTYPVLMAADV
MSMEG_1657|M.smegmatis_MC2_155 HSQLAWVLGCFTGFGQASRMTQFKDKSQKQGADATTVGLFTYPVLMAADV
TH_1974|M.thermoresistible__bu HAELAWVLGCFTGFGQASRMTQFKDKAQKQGTEGTTVGLFTYPVLMAADV
MLBr_00686|M.leprae_Br4923 HTQLAWVLGCFTGFGQASRMTQFKDKALRQGADSTTVGLFTYPVLQAADV
MAV_4310|M.avium_104 HTQLAWVLGCFTGFGQASRMTQFKDKSLRQGADSTTVGLFTYPVLQAADV
Mb3369c|M.bovis_AF2122/97 HTQLAWVLGCFTGFGQASRMTQFKDKSARQGSEATTVGLFTYPVLQAADV
Rv3336c|M.tuberculosis_H37Rv HTQLAWVLGCFTGFGQASRMTQFKDKSARQGSEATTVGLFTYPVLQAADV
MMAR_1188|M.marinum_M HSQLAWVLGCFTGFGQASRMTQFKDKSTRQGADSATVGLFTYPVLQAADV
MUL_1441|M.ulcerans_Agy99 HSQLAWVLGCFTGFGQASRMTQFKDKSTRQGADSATVGLFTYPVLQAADV
MAB_3683c|M.abscessus_ATCC_199 HAELAWVLGCLTGYGEAARMTQFKDKSAKQGNDATTVGLFTYPVLMAADI
*::*******:**:*:*:********: :** :.:********** ***:
Mflv_4871|M.gilvum_PYR-GCK LLYDTELVPVGEDQRQHLELARDVAQRINARFPDTFVVPEAMIPKATAKI
Mvan_1560|M.vanbaalenii_PYR-1 LLYDTGLVPVGEDQRQHLELARDLAQRINARFPGTFVVPEAMIPKATAKI
MSMEG_1657|M.smegmatis_MC2_155 LLYDTHVVPVGEDQRQHLELARDLAQRFNARFPDTFVVPEPMIPKATAKI
TH_1974|M.thermoresistible__bu LLYDTDLVPVGEDQRQHLELARDVAQRFNSRFPDTFVIPEAMIPRTAAKI
MLBr_00686|M.leprae_Br4923 LAYDTDLVPVGEDQRQHLELARDLAQRFNSRFPDTFVVPDMFIPKMAAKI
MAV_4310|M.avium_104 LAYDTGLVPVGEDQRQHLELARDVAQRFNARFPDTFVVPEMLIPKVTAKI
Mb3369c|M.bovis_AF2122/97 LAYDTELVPVGEDQRQHLELARDVAQRFNSRFPGTLVVPDVLIPKMTAKI
Rv3336c|M.tuberculosis_H37Rv LAYDTELVPVGEDQRQHLELARDVAQRFNSRFPGTLVVPDVLIPKMTAKI
MMAR_1188|M.marinum_M LAYDAELVPVGEDQRQHLELARDIAQRFNGRFPDTFVVPDVLIPKITAKI
MUL_1441|M.ulcerans_Agy99 LAYDAELVPVGEDQRQHLELARDIAQRFNGRFPDTFVVPDVLIPKITAKI
MAB_3683c|M.abscessus_ATCC_199 LLYQTNVVPVGDDQRQHLELTRDVAQRFNGRYGQTFVLPETFVPKNAARI
* *:: :****:********:**:***:*.*: *:*:*: ::*: :*:*
Mflv_4871|M.gilvum_PYR-GCK FDLQDPTAKMSKSAESDAGLISLLDDPAKTAKKIRSAVTDSEREIRFDRE
Mvan_1560|M.vanbaalenii_PYR-1 YDLQDPTAKMSKSADSDAGLISLLDDPAKTAKKIRSAVTDSEREIRFDRD
MSMEG_1657|M.smegmatis_MC2_155 YDLQDPTAKMSKSAATDAGLISLLDDPKATAKKIRSAVTDSEREIRFDPE
TH_1974|M.thermoresistible__bu YDLQDPTAKMSKSAGSDAGLIALLDDPKVTAKKIRSAVTDSEREIRYDPE
MLBr_00686|M.leprae_Br4923 YDLADPTSKMSKSASSDAGLINLLDDPALSVKKIRAAVTDSEREIRYDPE
MAV_4310|M.avium_104 YDLADPTSKMSKSASTDAGLVNLLDDPALSAKKIRSAVTDSEREIRFDTD
Mb3369c|M.bovis_AF2122/97 YDLQDPTSKMSKSAGTDAGLINLLDDPALSAKKIRSAVTDSERDIRYDPD
Rv3336c|M.tuberculosis_H37Rv YDLQDPTSKMSKSAGTDAGLINLLDDPALSAKKIRSAVTDSERDIRYDPD
MMAR_1188|M.marinum_M YDLQDPTSKMSKSASTDAGLISLLDAPAVSAKKIRSAVTDSEREIRYDPE
MUL_1441|M.ulcerans_Agy99 YDLQDPTSKMSKSASTDAGLISLLDAPAVSAKKIRSAVTDSEREIRYDPE
MAB_3683c|M.abscessus_ATCC_199 YDLQDPTVKMSKSASTPAGLINLLDEPAVSAKKVKSAQTDSDREIRFDRD
:** *** ****** : ***: *** * :.**:::* ***:*:**:* :
Mflv_4871|M.gilvum_PYR-GCK AKPGVSNLLTIQSAVTGTDIDKLVEGYAGRGYGDLKKETAEAVVEFVTPI
Mvan_1560|M.vanbaalenii_PYR-1 AKPGVSNLLTIQSAVTGVDVDKLVEGYAGRGYGDLKKETAEAVVEFVTPI
MSMEG_1657|M.smegmatis_MC2_155 AKPGISNLLTIQSAVTGTTVDKLVESYAGRGYGDLKKDTAEAVVEFVTPI
TH_1974|M.thermoresistible__bu AKPGVSNLLTIQSAVTGTDIDTLVARYAGRGYGDLKKDTAEAVVEFVTPI
MLBr_00686|M.leprae_Br4923 VKPGVSNLLNIQSAVTGVDVDTLVQRYVGHGYGDLKKDTAEAVVEFVSPI
MAV_4310|M.avium_104 AKPGVSNLLSIQSAVSGVDIDKLVQGYAGRGYGDLKKDTADAVVEFVTPI
Mb3369c|M.bovis_AF2122/97 VKPGVSNLLNIQSAVTGTDIDVLVDGYAGHGYGDLKKDTAEAVVEFVNPI
Rv3336c|M.tuberculosis_H37Rv VKPGVSNLLNIQSAVTGTDIDVLVDGYAGHGYGDLKKDTAEAVVEFVNPI
MMAR_1188|M.marinum_M AKPGVSNLLSIQSAVSRVSIDTLVDGYAGRGYGDLKKDTAEAVVAYVSPI
MUL_1441|M.ulcerans_Agy99 AKPGVSNLLSIQSAVSGVSIDTLVDGYAGRGYGDLKKDTAEAVVAYVSPI
MAB_3683c|M.abscessus_ATCC_199 AKPGVSNLLSIQSALTGADIETLVDGYAGRGYGDLKKDTAEALIEFVTPV
.***:****.****:: . :: ** *.*:*******:**:*:: :*.*:
Mflv_4871|M.gilvum_PYR-GCK KSTVDDLLGDPAELEAVLARGAERAREVSGQTLRRVYDRLGFLASR-
Mvan_1560|M.vanbaalenii_PYR-1 KARVDELLGDPAELEGVLARGAQRAREVSGVTLRRVYDRLGFLASR-
MSMEG_1657|M.smegmatis_MC2_155 KTRVDDLLSDRAELESVLAAGARRATEVSAKTLQRVYDRIGFLPQPS
TH_1974|M.thermoresistible__bu KTRVDELLADPAELEGILAAGAERAREVSATTLKRVYDRLGFLPPRS
MLBr_00686|M.leprae_Br4923 KDRVDELMADLTELEVVLAVGAQRAQDVAGKTMQRVYDRLGFLPQRG
MAV_4310|M.avium_104 KARVDELMADPAELEAVLAAGAQRAQDVAGKTVERVYDRLGFLPRRG
Mb3369c|M.bovis_AF2122/97 QARVDELTADPAELEAVLAAGAQRAHDVASKTVQRVYDRLGFLL---
Rv3336c|M.tuberculosis_H37Rv QARVDELTADPAELEAVLAAGAQRAHDVASKTVQRVYDRLGFLL---
MMAR_1188|M.marinum_M KARVDELTADLSELETVLAAGAERAHDIASKTVQRVYDRLGFLPQRG
MUL_1441|M.ulcerans_Agy99 KARVDELTADLSELETVLAAGAERAHDIASKTVQRVYDRLGFLPQRG
MAB_3683c|M.abscessus_ATCC_199 RDRTNELLSDTAALDDILASGAARARELAEKTLTEVYDRIGFVRPAR
: .::* .* : *: :** ** ** ::: *: .****:**: