For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MITHDAYPVEPWQVRETRLDLNLLAQSESLFALSNGHIGLRGNLEEGEPHGLPGTYLAGFFEMRPLPYAE AGFGYPEAGQTVVNVTNGKILRLLVDDEPFDVRYGELIDHERTLDMRAGTLNRRAHWCSPAGKQVRVETT RLVSLAHRGVAAIEYVVEAVDQFTRITVQSELVANEDQPESSDDPRVAAILKNPLQPVHHEKTDHGALLV HRTLASGLMVAAAMEHDVEVPGRVEVSTDAYEDLARTTVICGLRPGQKLRIVKYLAYGWSSLRSRPALRD QAAGAITGARYSGWEGLLESQRAYLDEFWDSADVEVEGDPDCQQAVRFGLFHVLQASARAERRAIAGKGL TGTGYDGHAFWDTEGFVLPVLTYTKPQAAADALRWRASTLDLARERAALLDLKGASFPWRTIRGEECSAY WPAGTAAWHINADIAMAFERYRIVTGDDSLEQECGLEVLVETARLWMSLGHHDRHGVWHLDGVTGPDEYT AVVRDNVFTNLMAAHNLRVAADACIRHPDAAYAMGVTTEETAAWRDAADAAHIPYDEELGVHPQCEGFTT LAEWDFSANTVYPLLMNEPYVRLYPAQVLKQADLVLAMQWQSHAFTPEQKARNVDYYERRTTRDSSLSAC TQAVMCADVGHLALAHDYTYEAAMIDLRDLHRNTRDGLHMASLAGAWTALVAGFGGLRDDEGILSLDPHL PDGISCLRFRLRWKGFRVTVQANHSDVTYTVRDGPDGTLAIRHAGEELVLSTSEPTTVAIRPRHPLLPVP PQPPGREPLRRRPAGH*
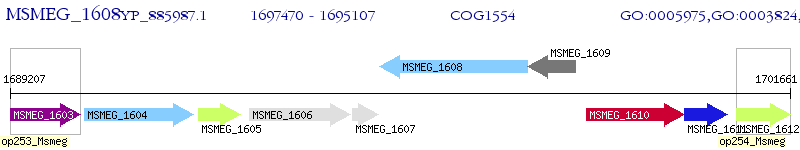
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1608 | - | - | 100% (787) | glycosyl transferase |
| M. smegmatis MC2 155 | MSMEG_3954 | - | 1e-54 | 28.70% (763) | trehalose 6-phosphate phosphorylase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3434 | - | 0.0 | 83.63% (782) | hypothetical protein Mb3434 |
| M. gilvum PYR-GCK | Mflv_1073 | - | 0.0 | 78.37% (786) | Kojibiose phosphorylase |
| M. tuberculosis H37Rv | Rv3401 | - | 0.0 | 83.76% (782) | hypothetical protein Rv3401 |
| M. leprae Br4923 | MLBr_00392 | - | 0.0 | 82.23% (782) | putative sugar phosphorylase |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1143 | - | 0.0 | 82.86% (782) | glycosyl hydrolase |
| M. avium 104 | MAV_4352 | - | 0.0 | 81.71% (782) | glycoside hydrolase family protein 65, central catalytic |
| M. thermoresistible (build 8) | TH_2141 | - | 0.0 | 80.33% (778) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0906 | - | 0.0 | 82.27% (626) | glycosyl hydrolase |
| M. vanbaalenii PYR-1 | Mvan_5750 | - | 0.0 | 79.13% (781) | Kojibiose phosphorylase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_1143|M.marinum_M -MISEEAFPVEPWQVRETRLDLNLLAQSESIFALSNGHIGLRGNLDEGEP
MUL_0906|M.ulcerans_Agy99 --------------------------------------------------
Mb3434|M.bovis_AF2122/97 -MITEDAFPVEPWQVRETKLNLNLLAQSESLFALSNGHIGLRGNLDEGEP
Rv3401|M.tuberculosis_H37Rv -MITEDAFPVEPWQVRETKLNLNLLAQSESLFALSNGHIGLRGNLDEGEP
MLBr_00392|M.leprae_Br4923 -MITEEAFPVEPWQIRETRLDLNLLAQSESLFALSNGHIGLRGNLDEGEP
MAV_4352|M.avium_104 -MIADESFPVDPWHVRETQLNLNLLAQSESLFTLSNGHIGLRGNLDEGEP
MSMEG_1608|M.smegmatis_MC2_155 MMITHDAYPVEPWQVRETRLDLNLLAQSESLFALSNGHIGLRGNLEEGEP
TH_2141|M.thermoresistible__bu VSPAEDIIPVEPWRVRETRLDFELLDETESLFALSNGHIGMRGNLDEGEP
Mflv_1073|M.gilvum_PYR-GCK MEMTDEDFPVEPWCIRETALRTELLAQTESLFALSNGHIGLRGNLDEGEP
Mvan_5750|M.vanbaalenii_PYR-1 -MIADEAFPVEPWHIRETELRLDLLPQTESLFALSNGHIGLRGNLDEGEP
MMAR_1143|M.marinum_M YGLPGTYLNSFFEIRPLPYAEAGYGYPEAGQTVVDVTNGKILRLLVDDEP
MUL_0906|M.ulcerans_Agy99 --------------------------------------------------
Mb3434|M.bovis_AF2122/97 FGLPGTYLNSFYEIRPLPYAEAGYGYPEAGQTVVDVTNGKIFRLLVGDEP
Rv3401|M.tuberculosis_H37Rv FGLPGTYLNSFYEIRPLPYAEAGYGYPEAGQTVVDVTNGKIFRLLVGDEP
MLBr_00392|M.leprae_Br4923 YGLPGTYLNSFYEIRPLPYAEAGYGYPEAGQTVVDVTNGKILRLFVEDEP
MAV_4352|M.avium_104 YGLPGTYLNSFYEIRPLPYAEAGYGYPEAGQTLVDVTNGKIIRLLVEDEP
MSMEG_1608|M.smegmatis_MC2_155 HGLPGTYLAGFFEMRPLPYAEAGFGYPEAGQTVVNVTNGKILRLLVDDEP
TH_2141|M.thermoresistible__bu HGLPGTYLNAFFEFRPLPYAEAGYGYPESGQSLINVTNGKIMRLLVDDEP
Mflv_1073|M.gilvum_PYR-GCK YSLPGTYLNGFYEIRPLPYAEAGFGYPEDGQSIVNVTNGKIIRLLVDDEP
Mvan_5750|M.vanbaalenii_PYR-1 HGLPGTYLNGFYEIRPLPYAEAGFGYPEEGQSIVDVTNGKLIRLLVDDEP
MMAR_1143|M.marinum_M FDVRYGELLSHERVLDLRAGTLTRSTHWRSPADKQVKVVSTRLVSLSHRS
MUL_0906|M.ulcerans_Agy99 --------------------------------------------------
Mb3434|M.bovis_AF2122/97 FDVRYGELISHERILDLRAGTLTRRAHWRSPAGKQVKVTSTRLVSLAHRS
Rv3401|M.tuberculosis_H37Rv FDVRYGELISHERILDLRAGTLTRRAHWRSPAGKQVKVTSTRLVSLAHRS
MLBr_00392|M.leprae_Br4923 FDVRYGEVIFHERVLDLCAGTLTRRANWRSPADKQVKVVSTRLVSLAHRS
MAV_4352|M.avium_104 FDVRYGELLSHERTLDLRGGTLTRSAYWRSPAGKEVKVVSTRLVSLAQRG
MSMEG_1608|M.smegmatis_MC2_155 FDVRYGELIDHERTLDMRAGTLNRRAHWCSPAGKQVRVETTRLVSLAHRG
TH_2141|M.thermoresistible__bu FDLRYGELISHERILDLRAGTLSRDARWCSPAGKQVRVRSTRVVSLAHRA
Mflv_1073|M.gilvum_PYR-GCK FDVRYGELHSHERVLDMRAGVLTRQVDWTAPSGTRVKVNSTRLVSLTQRG
Mvan_5750|M.vanbaalenii_PYR-1 FDVRYGELSSHERVLDLRAGTLTRAAEWTSPAGKRVEVHSTRLVSLTQRG
MMAR_1143|M.marinum_M VAAIEYTVEAVDDFARITVQSELVTNEDQPETSDDPRVSAILEHPLQAVD
MUL_0906|M.ulcerans_Agy99 -------MEAVDDFARITVQSELVTNEDQPETSDDPRVSAILEHPLQAVD
Mb3434|M.bovis_AF2122/97 VAAIEYVVEAIEEFVRVTVQSELVTNEDVPETSADPRVSAILDRPLQAVE
Rv3401|M.tuberculosis_H37Rv VAAIEYVVEAIEEFVRVTVQSELVTNEDVPETSADPRVSAILDRPLQAVE
MLBr_00392|M.leprae_Br4923 VAAIEYVVEALDKFVRVTVQSELVSNEDQPETSGDPRVSAILDNPLEAVE
MAV_4352|M.avium_104 VAAIEYVVQAVGDFVRVTVQSELVTNEDQPQTSGDPRVSAILENPLEEVD
MSMEG_1608|M.smegmatis_MC2_155 VAAIEYVVEAVDQFTRITVQSELVANEDQPESSDDPRVAAILKNPLQPVH
TH_2141|M.thermoresistible__bu VAAIEYIVEPVDEYALITVQSELVANEDIPHPEWDPRVAAALESPLEPVC
Mflv_1073|M.gilvum_PYR-GCK LAAIEYVVEVVDD-VLLTVQSELVANEDQPTPSDDPRVASALDDPLEAVQ
Mvan_5750|M.vanbaalenii_PYR-1 LAAIEYVVEAVDD-VLLTVQSELVANEDQPPASADPRVAAVLSHPLTPVQ
:: : . . :*******:*** * .. ****:: *. ** *
MMAR_1143|M.marinum_M HENTECGALLMHRTRSSALMMAAAMDHEVEVPGRVEITTDARPDLARTTV
MUL_0906|M.ulcerans_Agy99 HENTECGALLMHRTRSSALMMAAAMDHEVEVPGRVEITTDARPDLARTTV
Mb3434|M.bovis_AF2122/97 HERTERGALLMHRTRASALMMAAGMEHEVEVPGRVEITTDARPDLARTTV
Rv3401|M.tuberculosis_H37Rv HERTERGALLMHRTRASALMMAAGMEHEVEVPGRVEITTDARPDLARTTV
MLBr_00392|M.leprae_Br4923 HEATERGGLLMHRTRASSLMMAAGMDHEVEVPGRVEITTDARPDLARTTV
MAV_4352|M.avium_104 HENTDTGALLIHRTRSSALMMAAGMDHEVEVPGRVEVSTDSRADLARTTV
MSMEG_1608|M.smegmatis_MC2_155 HEKTDHGALLVHRTLASGLMVAAAMEHDVEVPGRVEVSTDAYEDLARTTV
TH_2141|M.thermoresistible__bu HETTDSGAVLIHRTRASELMMVAEMAHQVEVPGRMEMHTEAADDLARTTV
Mflv_1073|M.gilvum_PYR-GCK HELSERGAVLIHRTRTSELSMAAAMDHVVEADGRIDVSGDSGDDWARTTI
Mvan_5750|M.vanbaalenii_PYR-1 HEVSERGAMLIHRTRASELTLAAAMDHVVEAPGRVDLSGDAGQDWARTTV
** :: *.:*:*** :* * :.* * * **. **::: :: * ****:
MMAR_1143|M.marinum_M ICGLRPGQKLRIVKYLAYGWSSLRSRPALRDQVAGALHGARYSGWQGLID
MUL_0906|M.ulcerans_Agy99 ICGLRPGQKLRIVKYLAYGWSSLRSRPALRDQVAGALHGARYSGWQGLID
Mb3434|M.bovis_AF2122/97 ICGLRPGQKLRIVKYLAYGWSSLRSRPALRDQAAGALHGARYSGWQGLLD
Rv3401|M.tuberculosis_H37Rv ICGLRPGQKLRIVKYLAYGWSSLRSRPALRDQAAGALHGARYSGWQGLLD
MLBr_00392|M.leprae_Br4923 ICGLRPGQKLRIVKYLAYGWSSLRSRPALHDQATAALHTARYSGWQGLLD
MAV_4352|M.avium_104 ICGLRAGQKLRIVKYLAYGWSSMRSRPALRDQVAAALHSARYTGWPGLLE
MSMEG_1608|M.smegmatis_MC2_155 ICGLRPGQKLRIVKYLAYGWSSLRSRPALRDQAAGAITGARYSGWEGLLE
TH_2141|M.thermoresistible__bu ICGLRAGQRLRIVKWLAYGWSSLRSRPALRDQAAGAIAIARYTGWQGLLD
Mflv_1073|M.gilvum_PYR-GCK VCALKAGERLRLVKYLAYGWSSLRSRPALRDQVAAAIAGARYTGWQGLLD
Mvan_5750|M.vanbaalenii_PYR-1 VCGLKAGERLRLVKYLAYGWSSLRSRPALRDQVAAAIAGARYTGWDGLLT
:*.*:.*::**:**:*******:******:**.:.*: ***:** **:
MMAR_1143|M.marinum_M TQRSYLDEFWDSADVEVEGDPECQQAVRFGLFHLLQASARAERRAIPSKG
MUL_0906|M.ulcerans_Agy99 TQRSYLDEFWDSADVEVEGDPECQQAVRFGLFHLLQASARAERRAIPSKG
Mb3434|M.bovis_AF2122/97 AQRAYLDDFWDSADVEVEGDPECQQAVRFGLFHLLQASARAERRAIPSKG
Rv3401|M.tuberculosis_H37Rv AQRAYLDDFWDSADVEVEGDPECQQAVRFGLFHLLQASARAERRAIPSKG
MLBr_00392|M.leprae_Br4923 AQRAYLNEFWDSADVEVEGDPESQQAVRFGLFHLLQASARAERRAIPSKG
MAV_4352|M.avium_104 SQRKYLDNFWDCADVEVEGDPESQQAVRFGLFHLLQASARAERRAIAAKG
MSMEG_1608|M.smegmatis_MC2_155 SQRAYLDEFWDSADVEVEGDPDCQQAVRFGLFHVLQASARAERRAIAGKG
TH_2141|M.thermoresistible__bu AQRAYLDDFWDCADVEVEGDPDCQQAVRFGLFHVLQASARAERRAIPAKG
Mflv_1073|M.gilvum_PYR-GCK AQREYLDDYWDCADVEVEGDPDCQQAVRFGLFHVLQASARAERRAIAGKG
Mvan_5750|M.vanbaalenii_PYR-1 AQREYLDEFWDCADVEVDGDADCQQAVRFGLFHVLQASARAERRAIAGKG
:** **:::**.*****:**.:.**********:************..**
MMAR_1143|M.marinum_M LTGTGYDGHAFWDSEGFVLPVLTYTAPHAVADALRWRASTLDLAKERAAE
MUL_0906|M.ulcerans_Agy99 LTGTGYDGHAFWDSEGFVLPVLTYTAPHAVADALRWRASTLDLAKERAAE
Mb3434|M.bovis_AF2122/97 LTGTGYDGHAFWDTEGFVLPVLTYTAPHAVADALRWRASTLDLAKERAAE
Rv3401|M.tuberculosis_H37Rv LTGTGYDGHAFWDTEGFVLPVLTYTAPHAVADALRWRASTLDLAKERAAE
MLBr_00392|M.leprae_Br4923 LTGTGYDGHAFWDTEGFVLPVLTYTAPHAVADALRWRASTLQLAKDRAAE
MAV_4352|M.avium_104 LTGTGYDGHAFWDTEGYVLPVLTYTAPHTVADALRWRASTLDLAKERAAE
MSMEG_1608|M.smegmatis_MC2_155 LTGTGYDGHAFWDTEGFVLPVLTYTKPQAAADALRWRASTLDLARERAAL
TH_2141|M.thermoresistible__bu LTGTGYDGHAFWDTEGHVLPLLTYTAPYAAADALRWRASTLDMARERAAE
Mflv_1073|M.gilvum_PYR-GCK LTGTGYDGHAFWDTEGFVLPVLTYTAPRAAADALRWRASTLDKARERADE
Mvan_5750|M.vanbaalenii_PYR-1 LTGTGYDGHAFWDTEGFVLPVLTYTAPRAAADALRWRASTLEMARDRAAE
*************:**.***:**** * :.***********: *::**
MMAR_1143|M.marinum_M LGLDGASFPWRTIRGEECSAYWPAGTAAWHINADIAMAFERYRTVTGDHS
MUL_0906|M.ulcerans_Agy99 LGLDGASFPWRTIRGEECSAYWPAGTAAWQINADIAMAFERYRTVTCDHS
Mb3434|M.bovis_AF2122/97 LGLEGAAFPWRTIRGQESSAYWPAGTAAWHINADIAMAFERYRIVTGDGS
Rv3401|M.tuberculosis_H37Rv LGLEGAAFPWRTIRGQESSAYWPAGTAAWHINADIAMAFERYRIVTGDGS
MLBr_00392|M.leprae_Br4923 LGLDGASFCWRTIRGQECSAYWPAGTAAWHINADIAMAFERYRIVTGDHS
MAV_4352|M.avium_104 LGLEGAAFPWRTIRGQECSAYWPAGTAAWHINADIAMAFERYRIVTGDDD
MSMEG_1608|M.smegmatis_MC2_155 LDLKGASFPWRTIRGEECSAYWPAGTAAWHINADIAMAFERYRIVTGDDS
TH_2141|M.thermoresistible__bu LDLQGAAFPWRTIAGQECSAYWPAGTAGWHVNADIAMAFERYRIVTGDES
Mflv_1073|M.gilvum_PYR-GCK LDLKGASFPWRTIRGEECSAYWPAGTAAFHVNADIAMAFERYRVVTGDDS
Mvan_5750|M.vanbaalenii_PYR-1 LDLRGAAFPWRTIHGEECSAYWPAGTAGFHVNADIAMAFDRYRVVTGDES
*.* **:* **** *:*.*********.:::********:*** ** * .
MMAR_1143|M.marinum_M LEKDCGLAVLIETARLWISLGHHDRHGIWHLDGVTGPDEYTAIVRDNVFT
MUL_0906|M.ulcerans_Agy99 LEKDCGLAVLIETARLWISLGHHDRHGIWHLDGVTGPDEYTAIVRDNVFT
Mb3434|M.bovis_AF2122/97 LEEECGLAVLIETARLWLSLGHHDRHGVWHLDGVTGPDEYTAVVRDNVFT
Rv3401|M.tuberculosis_H37Rv LEEECGLAVLIETARLWLSLGHHDRHGVWHLDGVTGPDEYTAVVRDNVFT
MLBr_00392|M.leprae_Br4923 LEEECGLAVLIETARLWLSLGHHDRHGVWHLDGVTGPDEYTAVVRDNVFT
MAV_4352|M.avium_104 LEAECGLPVLVETARLWLSLGHHDRHGVWHLDGVTGPDEYTAVVRDNVFT
MSMEG_1608|M.smegmatis_MC2_155 LEQECGLEVLVETARLWMSLGHHDRHGVWHLDGVTGPDEYTAVVRDNVFT
TH_2141|M.thermoresistible__bu LEAECGLAVLVETARLWQSLGHHDRHGVWHLDGVTGPDEYTAVVRDNVFT
Mflv_1073|M.gilvum_PYR-GCK LEKECGLEVMVETARMWLSLGHHDRHGNWRIDGVTGPDEYTAVVRDNVFT
Mvan_5750|M.vanbaalenii_PYR-1 LEKDCGLAVLVDTARLWLSLGHHDRYGRWRIDGVTGPDEYTAVVRDNVFT
** :*** *:::***:* *******:* *::***********:*******
MMAR_1143|M.marinum_M NLMAAHNLITAADACLRHPEAAQDMGVTTEEMAAWRDAADAVNIPYDEEL
MUL_0906|M.ulcerans_Agy99 NLMAAHNLITAADACLRHPEAAQDMGVTTEGMAAWRDAADAVNIPYDEEL
Mb3434|M.bovis_AF2122/97 NLMAAHNLHTAADACLRHPEAAEAMGVTTEEMAAWRDAADAANIPYDEEL
Rv3401|M.tuberculosis_H37Rv NLMAAHNLHTAADACLRHPEAAEAMGVTTEEMAAWRDAADAANIPYDEEL
MLBr_00392|M.leprae_Br4923 NLMAASNLITAADACLRQPEAAKAMGVTTEEMAAWRDAADAANIPYDDEL
MAV_4352|M.avium_104 NLMAAHNLRVAAEACNRHPDGAKALGVTTEETAAWRDAADAVKIPYDSGL
MSMEG_1608|M.smegmatis_MC2_155 NLMAAHNLRVAADACIRHPDAAYAMGVTTEETAAWRDAADAAHIPYDEEL
TH_2141|M.thermoresistible__bu NLMAAANLRAAVAACNRHPDLARELGVTDEETEAWLAAADAVHIPYDEEL
Mflv_1073|M.gilvum_PYR-GCK NLMAAANLRNAADACTRHPELAEAMGVDSEETAAWRDAADAVHIPYDEEL
Mvan_5750|M.vanbaalenii_PYR-1 NLMAAANLRVAADACTRQPDAARALGVDTEETAAWRDAADAVHIPYDEEL
***** ** *. ** *:*: * :** * ** ****.:****. *
MMAR_1143|M.marinum_M GVHQQCEGFTTLAEWDFETNTTYPLLLHEAYVRLYPTQVIKQADLVLAMQ
MUL_0906|M.ulcerans_Agy99 GVHQQCEGFTTLAEWDFETNTTYPLLLHEAYVRLYPTQVIKQAHLVLAMQ
Mb3434|M.bovis_AF2122/97 GVHQQCEGFTTLAEWDFEANTTYPLLLHEAYVRLYPAQVIKQADLVLAMQ
Rv3401|M.tuberculosis_H37Rv GVHQQCEGFTTLAEWDFEANTTYPLLLHEAYVRLYPAQVIKQADLVLAMQ
MLBr_00392|M.leprae_Br4923 GVHQQCEGFTTFAEWDFEANTTYPLFLHEAYVRLYPAQVIKQADLVLAMQ
MAV_4352|M.avium_104 GVHQQCEGFTTFAEWDFEHNTTYPLLLHEAYVRLYPAQVIKQADLVLAMH
MSMEG_1608|M.smegmatis_MC2_155 GVHPQCEGFTTLAEWDFSANTVYPLLMNEPYVRLYPAQVLKQADLVLAMQ
TH_2141|M.thermoresistible__bu GVHQQCEGFTTLREWDFTTNTRYPLLKYEPYVRLYPSQVIKQADLVLAMH
Mflv_1073|M.gilvum_PYR-GCK GVHPQCEGFTTLREWDFHSNTEYPLLLNEPYVRLYPRQVVKQADLVLAMQ
Mvan_5750|M.vanbaalenii_PYR-1 GVHPQCDGFTTLREWDFDQNTKYPLLLHEPYVRLYPAQVVKQADLVLAMQ
*** **:****: **** ** ***: *.****** **:***.*****:
MMAR_1143|M.marinum_M WQSHAFTAEQKARNVDYYERRMVRDSSLSACTQAVMCADVGHLELAHDYA
MUL_0906|M.ulcerans_Agy99 WQSHAFTAEQKARNVDYYERRMVRDSSLSACTQAVMCADVGHLELAHDYA
Mb3434|M.bovis_AF2122/97 WQSHAFTPEQKARNVDYYERRMVRDSSLSACTQAVMCAEVGHLELAHDYA
Rv3401|M.tuberculosis_H37Rv WQSHAFTPEQKARNVDYYERRMVRDSSLSACTQAVMCAEVGHLELAHDYA
MLBr_00392|M.leprae_Br4923 WQSHAFTPEQKARNVDYYERRMVRDSSLSACTQAVMCAEVGHLELAHDYA
MAV_4352|M.avium_104 WQSHAFTPEQKARNVDYYEPRMVRDSSLSACTQAVMCAEVGHLELAHDYA
MSMEG_1608|M.smegmatis_MC2_155 WQSHAFTPEQKARNVDYYERRTTRDSSLSACTQAVMCADVGHLALAHDYT
TH_2141|M.thermoresistible__bu WQSHAFTPEQKARNVDYYERRTTRDSSLSAFTQAVMCAEVGHLELAHDYT
Mflv_1073|M.gilvum_PYR-GCK WQSHAFTDEQKARNVDYYERRTTRDSSLSACTQAVMCAEVGHLELGHDYA
Mvan_5750|M.vanbaalenii_PYR-1 WQSHAFTPDQKARNVDYYECRTTRDSSLSACTQAVMCAEVGHLELAHDYA
******* :********** * .******* *******:**** *.***:
MMAR_1143|M.marinum_M YEAALIDLRDLHRNTRDGLHMASLAGAWTALVGGFGGLRDDEGILSIDPQ
MUL_0906|M.ulcerans_Agy99 YEAALIDLRDLHRNTRDGLHMASLAGAWTALVGGFGGLRDDEGILSIDPQ
Mb3434|M.bovis_AF2122/97 YEAALIDLHDLHRNTRDGLHMASLAGAWTALVVGFGGLRDDEGILSIDPQ
Rv3401|M.tuberculosis_H37Rv YEAALIDLRDLHRNTRDGLHMASLAGAWTALVVGFGGLRDDEGILSIDPQ
MLBr_00392|M.leprae_Br4923 YEAALIDLRDLHRNTRDGLHMASLAGAWTALVGGFGGLRDDEGILSIDPQ
MAV_4352|M.avium_104 YEAALIDLRDLHHNTRDGLHMASLAGAWTALVSGFGGLRDDEGVLSLDPQ
MSMEG_1608|M.smegmatis_MC2_155 YEAAMIDLRDLHRNTRDGLHMASLAGAWTALVAGFGGLRDDEGILSLDPH
TH_2141|M.thermoresistible__bu YEAALVDLRDLHHNTRDGLHMASLAGAWTALVEGFGGLRDDDGVLQLDPA
Mflv_1073|M.gilvum_PYR-GCK YEAALIDLRNLHHNTGDGLHLASLAGAWTALVGGFGGLRDDEGVLSLDPQ
Mvan_5750|M.vanbaalenii_PYR-1 YEAALIDLRDLHHNTGDGLHLASLAGSWTALVAGFGGLRDDEGVLALDPQ
****::**::**:** ****:*****:***** ********:*:* :**
MMAR_1143|M.marinum_M LPDGISRLRFRLRWRGYRLTVDANHSTVTFTLRDGPGAELTIRHAGKDLE
MUL_0906|M.ulcerans_Agy99 LPDGISRLRFRLRWRGYRLTVDANHSTVTFTLRDGPGAELTIRHAGKDLE
Mb3434|M.bovis_AF2122/97 LPDGISRLRFRLRWRGFRLIVDANHTDVTFILGDGPGTQLTMRHAGQDLT
Rv3401|M.tuberculosis_H37Rv LPDGISRLRFRLRWRGFRLIVDANHTDVTFILGDGPGTQLTMRHAGQDLT
MLBr_00392|M.leprae_Br4923 LPHGISRLRFRLRWRGFRLTVDARHADVTYTLRDGPGGELTIRHAGEHLT
MAV_4352|M.avium_104 LPDGISRLRFRLRWRDFRVTVDVNHADVTYTLRDGPGGELTIQHAGAELE
MSMEG_1608|M.smegmatis_MC2_155 LPDGISCLRFRLRWKGFRVTVQANHSDVTYTVRDGPDGTLAIRHAGEELV
TH_2141|M.thermoresistible__bu LPSGITRLRFRLRWRGFRVKVDITHEQITYTLRDGPYGELTIRHAGEPLK
Mflv_1073|M.gilvum_PYR-GCK LPTGITRLRFRLRWRGFRVTVEADHDTVTYTLRDGPHGALEIRHAGEPLD
Mvan_5750|M.vanbaalenii_PYR-1 LPGGISRLRFRLRWRGFRVTVDADHDAVTYTLRDGPEGVLTIRHSGDPLE
** **: *******:.:*: *: * :*: : *** * ::*:* *
MMAR_1143|M.marinum_M LSSTKPTTISVHKHKPLLPPPPQPPGREPVHRRNMAAKVWASTAT-
MUL_0906|M.ulcerans_Agy99 LSSTKPTTISVHKHKPLLPPPPQPPGREPVHRRNMAAKVWASTAT-
Mb3434|M.bovis_AF2122/97 LHTDTPSTIAVRTRKPLLPPPPQPPGREPVHRR------ALAR---
Rv3401|M.tuberculosis_H37Rv LHTDTPSTIAVRTRKPLLPPPPQPPGREPVHRR------ALAR---
MLBr_00392|M.leprae_Br4923 LKSDSPSTIAVRDRKPLLPPPSQPPGREPVSRRHKSLIISAAR---
MAV_4352|M.avium_104 LSTQSPTTVAVRPRKPLLPPPQQPPGREPTHRRAVSRSSPGQ----
MSMEG_1608|M.smegmatis_MC2_155 LSTSEPTTVAIRPRHPLLPVPPQPPGREPLRRR------PAGH---
TH_2141|M.thermoresistible__bu LRTTEPTVVPMRPRKALLPPPPQPPGREPIRRRMLTTGAHRTA---
Mflv_1073|M.gilvum_PYR-GCK ITTDDATRVAVRARKALLPTPQQPPGREPQLRRR-RADLKR-----
Mvan_5750|M.vanbaalenii_PYR-1 INTRKPTRVAVRPKKPLLAPPKQPPGREPLRRWRSTVDASRNSRES
: : .: :.:: ::.**. * ******* *