For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTRTVLTIDTATPAVSAGVVRLSDGEAPATLAERVTIDARAHAEQLTPNIVAALGDAGITAGDLDAVVVG CGPGPFTGLRVGMASAAAFAHAIGVPVHGVCSLDAIGGDTRGDVLVVTDARRREVYWARYHDGARVAGPS VNAPADVPALLDAPVAAVAGSPAHTALFDLPVLEQVYPTPAGLVAAVADWDDPQPLVPLYLRRPDAKPSA AVKR
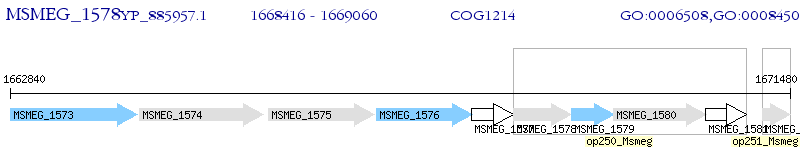
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1578 | - | - | 100% (214) | peptidase M22, glycoprotease |
| M. smegmatis MC2 155 | MSMEG_1580 | - | 3e-08 | 32.80% (125) | O-sialoglycoprotein endopeptidase |
| M. smegmatis MC2 155 | MSMEG_5923 | - | 6e-05 | 28.30% (159) | acetyl-CoA acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3455c | - | 6e-78 | 71.01% (207) | hypothetical protein Mb3455c |
| M. gilvum PYR-GCK | Mflv_4931 | - | 6e-89 | 77.67% (215) | peptidase M22, glycoprotease |
| M. tuberculosis H37Rv | Rv3421c | - | 6e-78 | 71.01% (207) | hypothetical protein Rv3421c |
| M. leprae Br4923 | MLBr_00378 | - | 7e-76 | 67.79% (208) | putative acetyltransferase |
| M. abscessus ATCC 19977 | MAB_3736c | - | 3e-65 | 62.25% (204) | hypothetical protein MAB_3736c |
| M. marinum M | MMAR_1122 | - | 2e-75 | 68.60% (207) | hypothetical protein MMAR_1122 |
| M. avium 104 | MAV_4369 | - | 3e-76 | 68.27% (208) | peptidase M22, glycoprotease |
| M. thermoresistible (build 8) | TH_1691 | - | 3e-75 | 66.97% (218) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0882 | - | 1e-75 | 68.60% (207) | hypothetical protein MUL_0882 |
| M. vanbaalenii PYR-1 | Mvan_1487 | - | 8e-84 | 73.49% (215) | peptidase M22, glycoprotease |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4931|M.gilvum_PYR-GCK ----MSRLVLAVDTATPAVTAGIVRV-DGDAIEVLAEQVTVDARAHAEQL
Mvan_1487|M.vanbaalenii_PYR-1 ----MSRLVLAIDTATPAVTAGVLRV-DGDAVEVLAEQVTVDARAHAERL
MSMEG_1578|M.smegmatis_MC2_155 ----MTRTVLTIDTATPAVSAGVVRLSDGEAPATLAERVTIDARAHAEQL
Mb3455c|M.bovis_AF2122/97 MSRVQISTVLAIDTATPAVTAGIVRR-H--DLVVLGERVTVDARAHAERL
Rv3421c|M.tuberculosis_H37Rv MSRVQISTVLAIDTATPAVTAGIVRR-H--DLVVLGERVTVDARAHAERL
MMAR_1122|M.marinum_M -----MTIILALDTSTPAVTAGVVQL-TGSECVALAERVTVDARAHAERL
MUL_0882|M.ulcerans_Agy99 -----MTIILALDTSTPAVTAGVVQL-TGSECVALAERVTVDARAHAERL
MAV_4369|M.avium_104 ----MSPLVLALDTSTPAVTAGIVRR-D--DLSVLAQRITVDARAHAERL
MLBr_00378|M.leprae_Br4923 -----MSIVLAIDTATAAVTAGIVAF-DGHDCFTLAERVTVDAKAHVERL
TH_1691|M.thermoresistible__bu ----MTMLILTIDTATPAVSAGVVRR-TPAGEELLAEKVTVDARAHAERL
MAB_3736c|M.abscessus_ATCC_199 -----MTLILALDTATAAITAGLVRRAPDGSVQPLAERITMGAKGHAEAL
:*::**:*.*::**:: *.:::*:.*:.*.* *
Mflv_4931|M.gilvum_PYR-GCK TPNIVAALGDAGIDASRLDAVVVGCGPGPFTGLRVGMATAAAFGHALGVP
Mvan_1487|M.vanbaalenii_PYR-1 TPNIVDALADAGVSVGQLDAVVVGCGPGPFTGLRVGMATAAAFGHALGVP
MSMEG_1578|M.smegmatis_MC2_155 TPNIVAALGDAGITAGDLDAVVVGCGPGPFTGLRVGMASAAAFAHAIGVP
Mb3455c|M.bovis_AF2122/97 TPNVLAALADAALTMADLDAVVVGCGPGPFTGLRAGMASAAAYGHALGIP
Rv3421c|M.tuberculosis_H37Rv TPNVLAALADAALTMADLDAVVVGCGPGPFTGLRAGMASAAAYGHALGIP
MMAR_1122|M.marinum_M TPNVLAALAGAALTMADLNAVVVGCGPGPFTGLRAGMATGAAYGHALDIP
MUL_0882|M.ulcerans_Agy99 TPNVLAALAGAALTMADLNAVVVGCGPGPFTGLRAGMATGAAYGHALDIP
MAV_4369|M.avium_104 TPNVLAALRDSGLNMTDLAAVVVGCGPGPFTGLRAGMATAAAYGHALGIP
MLBr_00378|M.leprae_Br4923 TPNVLVALADAELAMCELDAVVVGCGPGPFTGLRVGMATAAAYGHALGIP
TH_1691|M.thermoresistible__bu TPNVVAALAEAGLAFDDLDAVVVGCGPGPFTGLRVGMASAAAYGHALDIP
MAB_3736c|M.abscessus_ATCC_199 TPNIGVACADAGVAVGDLGAIVVGCGPGPFTGLRVGMATAAAMGLALDIP
***: * : : * *:*************.***:.** . *:.:*
Mflv_4931|M.gilvum_PYR-GCK VHGVCSLDAIA---AGTTGDVLVVTDARRREVYWARYRDGRRVDGPAVNS
Mvan_1487|M.vanbaalenii_PYR-1 VRGVCSLDAIA---AGSTGDVLVVTDARRREVYWARYRDGLRVDGPAVNA
MSMEG_1578|M.smegmatis_MC2_155 VHGVCSLDAIG---GDTRGDVLVVTDARRREVYWARYHDGARVAGPSVNA
Mb3455c|M.bovis_AF2122/97 VYGVCSLDAIG---GQTIGDTLVVTDARRREVYWARYCDGIRTVGPAVNA
Rv3421c|M.tuberculosis_H37Rv VYGVCSLDAIG---GQTIGDTLVVTDARRREVYWARYCDGIRTVGPAVNA
MMAR_1122|M.marinum_M VYGVCSLDAIG---GLTTGDTLVVTDARRREVYWARYRDGVRTHGPAVNA
MUL_0882|M.ulcerans_Agy99 VYGVCSLDAIG---GLTTGDTLVVTDARRREVYWARYRDGVRTHGPAVNA
MAV_4369|M.avium_104 VHGVCSLDAIG---GQTTGDTLVVTGARRREVYWARYRDGVRTDGPAVAA
MLBr_00378|M.leprae_Br4923 VHGVCSLDAIG---VRTTGDTLVVTDARRHEVYWARYRDGVRIAGPAVGS
TH_1691|M.thermoresistible__bu VHGVCSLDAIGVDTADTAGHVLVVTDARRREVYWARYRGGVRTDGPAVDS
MAB_3736c|M.abscessus_ATCC_199 VYPVCTLDAIG---HGTAGRVLVVTDARRREVYWAGYSDGVRVSGPAVDA
* **:****. : * .****.***:***** * .* * **:* :
Mflv_4931|M.gilvum_PYR-GCK AVDVP----AGAAAVAGSPEHTALFELPRLAPVYPSAAGLVAAVRDWDTD
Mvan_1487|M.vanbaalenii_PYR-1 AVDVP----GGADAVAGSPEHAALFELPRLAPVYPTAAGLVAAVADWVSE
MSMEG_1578|M.smegmatis_MC2_155 PADVPALLDAPVAAVAGSPAHTALFDLPVLEQVYPTPAGLVAAVADWD-D
Mb3455c|M.bovis_AF2122/97 AADVDP---GPALAVAGAPEHAALFALPCVEPSRPSPAGLVAAVN-WADK
Rv3421c|M.tuberculosis_H37Rv AADVDP---GPALAVAGAPEHAALFALPCVEPSRPSPAGLVAAVN-WADK
MMAR_1122|M.marinum_M PGDVDP---GTAHAVAGSPEHAGLFDLPRCEPSYPTVAGLAAAVN-WAQD
MUL_0882|M.ulcerans_Agy99 PGDVDP---GTAHAVAGSPEHAGLFDLPRCEPSYPTVAGLAAAVN-WAQD
MAV_4369|M.avium_104 PADVDP---GGAEAVAGSPEHARLFGLPVTGPAHPTPAGLVAAVGDWSVR
MLBr_00378|M.leprae_Br4923 PTDVDP---GTALTVAGSPEHAALFGLPLCEPIYPTPAGLVAAVPDWSVS
TH_1691|M.thermoresistible__bu PADVPT-----ADAVAGSRDHIGLFDLPALPPARPTPVGLVRVVPDWDSP
MAB_3736c|M.abscessus_ATCC_199 PADVSL---DGYTQVAGSPDHTTLFDLPAVDRHYPSPSRLVQVAG-WDEP
. ** ***: * ** ** *: *. .. *
Mflv_4931|M.gilvum_PYR-GCK PAPLVPLYLRRPDAKPSQAVKR----------------------------
Mvan_1487|M.vanbaalenii_PYR-1 PDPLVPLYLRRPDAKPPRAVAR----------------------------
MSMEG_1578|M.smegmatis_MC2_155 PQPLVPLYLRRPDAKPSAAVKR----------------------------
Mb3455c|M.bovis_AF2122/97 PAPLVPLYLRRPDAKPLAVCT-----------------------------
Rv3421c|M.tuberculosis_H37Rv PAPLVPLYLRRPDAKPLAVCT-----------------------------
MMAR_1122|M.marinum_M PAPLVPLYLRRPDAKPLAARP-----------------------------
MUL_0882|M.ulcerans_Agy99 PAPLVPLYLRRPDAKPLAARP-----------------------------
MAV_4369|M.avium_104 PQPLVPLYLRRPDATPRAARP-----------------------------
MLBr_00378|M.leprae_Br4923 PIPLVALYLRRPDAKPITADNEPIVIGTLTPADVDRCAQLESQFFDGDNP
TH_1691|M.thermoresistible__bu PAALKPLYLRRPDAKPAVARNR----------------------------
MAB_3736c|M.abscessus_ATCC_199 PGALTPLYLRRPDAKEPRR-------------------------------
* .* .********.
Mflv_4931|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1487|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1578|M.smegmatis_MC2_155 --------------------------------------------------
Mb3455c|M.bovis_AF2122/97 --------------------------------------------------
Rv3421c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1122|M.marinum_M --------------------------------------------------
MUL_0882|M.ulcerans_Agy99 --------------------------------------------------
MAV_4369|M.avium_104 --------------------------------------------------
MLBr_00378|M.leprae_Br4923 WPAAAFDRELANSYNCYVGARTADTLVGYAGITRLGHTPPFEYEVHTIAV
TH_1691|M.thermoresistible__bu --------------------------------------------------
MAB_3736c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4931|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1487|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_1578|M.smegmatis_MC2_155 --------------------------------------------------
Mb3455c|M.bovis_AF2122/97 --------------------------------------------------
Rv3421c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1122|M.marinum_M --------------------------------------------------
MUL_0882|M.ulcerans_Agy99 --------------------------------------------------
MAV_4369|M.avium_104 --------------------------------------------------
MLBr_00378|M.leprae_Br4923 DPAYRGRGVGRRLLGELLDFAGSGAIYLEVRTDNETAIALYRSVGFERIG
TH_1691|M.thermoresistible__bu --------------------------------------------------
MAB_3736c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4931|M.gilvum_PYR-GCK ---------------------
Mvan_1487|M.vanbaalenii_PYR-1 ---------------------
MSMEG_1578|M.smegmatis_MC2_155 ---------------------
Mb3455c|M.bovis_AF2122/97 ---------------------
Rv3421c|M.tuberculosis_H37Rv ---------------------
MMAR_1122|M.marinum_M ---------------------
MUL_0882|M.ulcerans_Agy99 ---------------------
MAV_4369|M.avium_104 ---------------------
MLBr_00378|M.leprae_Br4923 LRPRYYPASGADAYLMRREAQ
TH_1691|M.thermoresistible__bu ---------------------
MAB_3736c|M.abscessus_ATCC_199 ---------------------