For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTARNLGTEAKRQSERTKLLEERPVNLDGFVQEWPEVGMVAMDSAFDPEPSVRVENGVIVEMDGRARAD FDFIDQFIADHAIDVATTEQSMALPAVEIARMLVDPRVTRDEVIAVTGGLTPAKLLEVAKNLNIVEIMMG IQKMRARRTPANQAHCTSARDNPLQVACEAAEASLRGFSEVETTLGVVRYAPLVAMALQIGSQVGSGGLL TQCALEEATELELGMRGITAYAETISVYGTESVFVDGDDTPWSKAFLAAAYASRGIKMRFTSGTGSEVQM GNAEGRSMLYLEIRCILVAKGAGVQGLQNGSISCIGVPGAVPAGIRAVAAENLIASAVDLECASGNDQSF SHSPMRRTARLLPQMLPGTDFVCSGYSAVPNYDNMFAGSNLDAEDFDDFNTIQRDLQVDGGLRHVNEAEI VAARRRAAQALQAVFRYLDLPAITDAEIEAAVYAHGSRELIPRDVLEDLKGAQQVMDRNVTGLDLVKALE STGFADIAENLLAVLRQRVSGDLLQTSAIMTRDLKPLSAVNDRNDYAGPGTGYRPSGARWEEMKRLRHVT SAENPELEVE
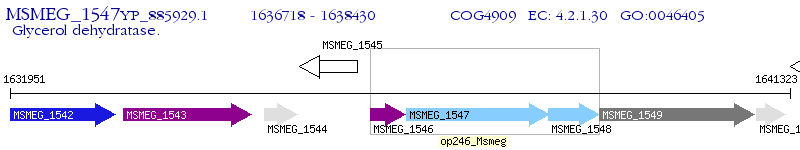
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1547 | - | - | 100% (570) | glycerol dehydratase large subunit |
| M. smegmatis MC2 155 | MSMEG_0497 | - | e-155 | 50.90% (554) | glycerol dehydratase large subunit |
| M. smegmatis MC2 155 | MSMEG_6321 | - | e-149 | 50.55% (546) | glycerol dehydratase large subunit |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1245 | - | 1e-145 | 48.92% (554) | glycerol dehydratase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5563 | - | 1e-148 | 50.37% (546) | glycerol dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1245|M.gilvum_PYR-GCK --MADELG--------RFRVLNSKPVNLDGFSVPDAALGLVAMSSPHDPA
Mvan_5563|M.vanbaalenii_PYR-1 --MADELG--------RFRVLNSKPVNLDGFSVPDAGLGLVAMSSPHDPA
MSMEG_1547|M.smegmatis_MC2_155 MTTARNLGTEAKRQSERTKLLEERPVNLDGFVQEWPEVGMVAMDSAFDPE
* :** * ::*:.:******* . :*:***.*..**
Mflv_1245|M.gilvum_PYR-GCK PSLVIRDGAVVELDSKDVAEFDVIDEFIARYGIDLSVAEEAMALDDETLA
Mvan_5563|M.vanbaalenii_PYR-1 PSLKIRGGEVVELDGKGAGEFDVIDEFIARYGIDLTVAEEAMALGDETLA
MSMEG_1547|M.smegmatis_MC2_155 PSVRVENGVIVEMDGRARADFDFIDQFIADHAIDVATTEQSMALPAVEIA
**: :..* :**:*.: .:**.**:*** :.**::.:*::*** :*
Mflv_1245|M.gilvum_PYR-GCK RMVVDINVPRAGVVRLIGGTTPAKLARVVALLSPVEMQMAMVKMRARRTP
Mvan_5563|M.vanbaalenii_PYR-1 RMVVDINVPRAEVVRLIGGTTPAKLARVVALLSPVEMQMAMAKMRARRTP
MSMEG_1547|M.smegmatis_MC2_155 RMLVDPRVTRDEVIAVTGGLTPAKLLEVAKNLNIVEIMMGIQKMRARRTP
**:** .*.* *: : ** ***** .*. *. **: *.: ********
Mflv_1245|M.gilvum_PYR-GCK SNQAHVTNQLDDPLLIAADAASAVAYGFREVETTVPVLGDAPSNAVALLI
Mvan_5563|M.vanbaalenii_PYR-1 SNQAHVTNQLDDPLLIAADAASAVAYGFREVETTVPVLGDAPSNAVALLI
MSMEG_1547|M.smegmatis_MC2_155 ANQAHCTSARDNPLQVACEAAEASLRGFSEVETTLGVVRYAPLVAMALQI
:**** *. *:** :*.:**.* ** *****: *: ** *:** *
Mflv_1245|M.gilvum_PYR-GCK GSQVGTPGAMAQCSIEEALELRLGLRGLTSYAETISIYGTEQVFIDGDDT
Mvan_5563|M.vanbaalenii_PYR-1 GSQVGSPGAMAQCSIEEALELRLGLRGLTSYAETISIYGTEQVFVDGDDT
MSMEG_1547|M.smegmatis_MC2_155 GSQVGSGGLLTQCALEEATELELGMRGITAYAETISVYGTESVFVDGDDT
*****: * ::**::*** **.**:**:*:******:****.**:*****
Mflv_1245|M.gilvum_PYR-GCK PFSKAILTSAYASRGLKMRVTSGGGAEVLMGAAQKCSILYLESRCVSLAR
Mvan_5563|M.vanbaalenii_PYR-1 PFSKAILTSAYASRGLKMRVTSGGGAEVLMGAAEKCSILYLESRCVSLAR
MSMEG_1547|M.smegmatis_MC2_155 PWSKAFLAAAYASRGIKMRFTSGTGSEVQMGNAEGRSMLYLEIRCILVAK
*:***:*::******:***.*** *:** ** *: *:**** **: :*:
Mflv_1245|M.gilvum_PYR-GCK ALGSQGVQNGGIDGVGVVASVPEGMKELLAENLMVMMRDLESCAGNDNLI
Mvan_5563|M.vanbaalenii_PYR-1 ALGSQGVQNGGIDGVGVVASVPEGMKELLAENLMVMMRDLESCAGNDNLI
MSMEG_1547|M.smegmatis_MC2_155 GAGVQGLQNGSISCIGVPGAVPAGIRAVAAENLIASAVDLECASGNDQSF
. * **:***.*. :** .:** *:: : ****:. ***..:***: :
Mflv_1245|M.gilvum_PYR-GCK SESDIRRSAHTLPVLLAGADFIFSGFGSIPRYDNAFALSNFNSDDMDDFL
Mvan_5563|M.vanbaalenii_PYR-1 SESDIRRSAHTLPVLLAGADFVFSGFGSIPRYDNAFALSNFNSDDMDDFL
MSMEG_1547|M.smegmatis_MC2_155 SHSPMRRTARLLPQMLPGTDFVCSGYSAVPNYDNMFAGSNLDAEDFDDFN
*.* :**:*: ** :*.*:**: **:.::*.*** ** **::::*:***
Mflv_1245|M.gilvum_PYR-GCK VLQRDWGADGGLRTVSPEHLEAVRRRAAQAVQAVYRDLGLADYDDARVED
Mvan_5563|M.vanbaalenii_PYR-1 VLQRDWGADGGLRTVSPEHLEAVRRRAAKAVQAVYRDLGLADYEDARVEE
MSMEG_1547|M.smegmatis_MC2_155 TIQRDLQVDGGLRHVNEAEIVAARRRAAQALQAVFRYLDLPAITDAEIEA
.:*** .***** *. .: *.*****:*:***:* *.*. **.:*
Mflv_1245|M.gilvum_PYR-GCK VVAANGSRDLPAGHPKMVAEAAASIEARQLTVFDVIAALHRTGFTEEAEA
Mvan_5563|M.vanbaalenii_PYR-1 VVAANGSRDLPAGHPKMVAEAAASIEARQLTVFDVIASLHRTGFTDEAEA
MSMEG_1547|M.smegmatis_MC2_155 AVYAHGSRELIPRDVLEDLKGAQQVMDRNVTGLDLVKALESTGFADIAEN
.* *:***:* . . :.* .: *::* :*:: :*. ***:: **
Mflv_1245|M.gilvum_PYR-GCK ITTLTRERLRGDQLQTSAIFDEQFRVFSKLTDPNDYRGPATGYALTEQRR
Mvan_5563|M.vanbaalenii_PYR-1 ITTLTRERLRGDQLQTSAIFDEKFRVLSKLTDPNDYTGPATGYALTDRRR
MSMEG_1547|M.smegmatis_MC2_155 LLAVLRQRVSGDLLQTSAIMTRDLKPLSAVNDRNDYAGPGTGYRPSGARW
: :: *:*: ** ******: ..:: :* :.* *** **.*** : *
Mflv_1245|M.gilvum_PYR-GCK AEIDAIRQARSGAELTVDQEEHRGHVVVTDVEPAHQGSDPREVCIGLSPA
Mvan_5563|M.vanbaalenii_PYR-1 AEIDAIRQARSSAELTADQESYRGHVLVTDVEPAQQGSDPREVCIGLSPA
MSMEG_1547|M.smegmatis_MC2_155 EEMKRLRHVTSAENPELEVE------------------------------
*:. :*:. *. : : *
Mflv_1245|M.gilvum_PYR-GCK WGRSVWLTLCGLTVGEVLRQIAAGLEEEGCIARTVRVCSTIDTGLIGLTA
Mvan_5563|M.vanbaalenii_PYR-1 WGRSVWLTLCGLTIGEVLRQISAGLEEEGCIARPVRVRSTIDVGLIGLTA
MSMEG_1547|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1245|M.gilvum_PYR-GCK ARLSGSGIGIGLQGKGTALIHRRDLAPLANLELFSVAPLLTAKMYRELGK
Mvan_5563|M.vanbaalenii_PYR-1 ARLSGSGIGIGLQGKGTALIHRRDLAPLANLELFSVAPLLTAKMYRELGK
MSMEG_1547|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1245|M.gilvum_PYR-GCK NAARHAKGMAPVPIFTGGTDESISARYHARAVALVALERDACEPGRAPVT
Mvan_5563|M.vanbaalenii_PYR-1 NAARHAKGMAPVPIFTGGTDESISARYHARAVALVALERESCEPGQPPVT
MSMEG_1547|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1245|M.gilvum_PYR-GCK VKVEAR
Mvan_5563|M.vanbaalenii_PYR-1 VKVEWP
MSMEG_1547|M.smegmatis_MC2_155 ------