For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VADELGRFRVLNSKPVNLDGFSVPDAALGLVAMSSPHDPAPSLVIRDGAVVELDSKDVAEFDVIDEFIAR YGIDLSVAEEAMALDDETLARMVVDINVPRAGVVRLIGGTTPAKLARVVALLSPVEMQMAMVKMRARRTP SNQAHVTNQLDDPLLIAADAASAVAYGFREVETTVPVLGDAPSNAVALLIGSQVGTPGAMAQCSIEEALE LRLGLRGLTSYAETISIYGTEQVFIDGDDTPFSKAILTSAYASRGLKMRVTSGGGAEVLMGAAQKCSILY LESRCVSLARALGSQGVQNGGIDGVGVVASVPEGMKELLAENLMVMMRDLESCAGNDNLISESDIRRSAH TLPVLLAGADFIFSGFGSIPRYDNAFALSNFNSDDMDDFLVLQRDWGADGGLRTVSPEHLEAVRRRAAQA VQAVYRDLGLADYDDARVEDVVAANGSRDLPAGHPKMVAEAAASIEARQLTVFDVIAALHRTGFTEEAEA ITTLTRERLRGDQLQTSAIFDEQFRVFSKLTDPNDYRGPATGYALTEQRRAEIDAIRQARSGAELTVDQE EHRGHVVVTDVEPAHQGSDPREVCIGLSPAWGRSVWLTLCGLTVGEVLRQIAAGLEEEGCIARTVRVCST IDTGLIGLTAARLSGSGIGIGLQGKGTALIHRRDLAPLANLELFSVAPLLTAKMYRELGKNAARHAKGMA PVPIFTGGTDESISARYHARAVALVALERDACEPGRAPVTVKVEAR
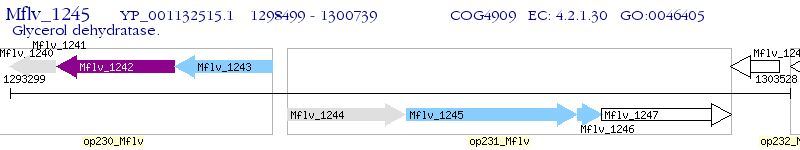
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1245 | - | - | 100% (746) | glycerol dehydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_6321 | - | 0.0 | 86.35% (740) | glycerol dehydratase large subunit |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5563 | - | 0.0 | 94.35% (744) | glycerol dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1245|M.gilvum_PYR-GCK -----------MADELGRFRVLNSKPVNLDGFSVPDAALGLVAMSSPHDP
Mvan_5563|M.vanbaalenii_PYR-1 -----------MADELGRFRVLNSKPVNLDGFSVPDAGLGLVAMSSPHDP
MSMEG_6321|M.smegmatis_MC2_155 MVAVTESGDPGQSPELGRMRILDAKPVNLDGFSVPDPDLGLAAMSSPHDP
: ****:*:*::************. ***.********
Mflv_1245|M.gilvum_PYR-GCK APSLVIRDGAVVELDSKDVAEFDVIDEFIARYGIDLSVAEEAMALDDETL
Mvan_5563|M.vanbaalenii_PYR-1 APSLKIRGGEVVELDGKGAGEFDVIDEFIARYGIDLTVAEEAMALGDETL
MSMEG_6321|M.smegmatis_MC2_155 QPSLVIRDGRVVEMDGKAAEDFDVIDEFIARYGLDLDVAPEAMAMSDIDL
*** **.* ***:*.* . :************:** ** ****:.* *
Mflv_1245|M.gilvum_PYR-GCK ARMVVDINVPRAGVVRLIGGTTPAKLARVVALLSPVEMQMAMVKMRARRT
Mvan_5563|M.vanbaalenii_PYR-1 ARMVVDINVPRAEVVRLIGGTTPAKLARVVALLSPVEMQMAMAKMRARRT
MSMEG_6321|M.smegmatis_MC2_155 ARMAVDINVPRAEVVRLIAGTTPAKLAKVIAVLTPVEMQAAMAKMRARRT
***.******** *****.********:*:*:*:***** **.*******
Mflv_1245|M.gilvum_PYR-GCK PSNQAHVTNQLDDPLLIAADAASAVAYGFREVETTVPVLGDAPSNAVALL
Mvan_5563|M.vanbaalenii_PYR-1 PSNQAHVTNQLDDPLLIAADAASAVAYGFREVETTVPVLGDAPSNAVALL
MSMEG_6321|M.smegmatis_MC2_155 PSNQAHVTNQLDDPLLIAADAASAVAYGFREVETTVPVLADAPSNAVALL
***************************************.**********
Mflv_1245|M.gilvum_PYR-GCK IGSQVGTPGAMAQCSIEEALELRLGLRGLTSYAETISIYGTEQVFIDGDD
Mvan_5563|M.vanbaalenii_PYR-1 IGSQVGSPGAMAQCSIEEALELRLGLRGLTSYAETISIYGTEQVFVDGDD
MSMEG_6321|M.smegmatis_MC2_155 IGSQVGVPGAMAQCSIEEALELRLGLRGLTSYAETISIYGTEQVFVDGDD
****** **************************************:****
Mflv_1245|M.gilvum_PYR-GCK TPFSKAILTSAYASRGLKMRVTSGGGAEVLMGAAQKCSILYLESRCVSLA
Mvan_5563|M.vanbaalenii_PYR-1 TPFSKAILTSAYASRGLKMRVTSGGGAEVLMGAAEKCSILYLESRCVSLA
MSMEG_6321|M.smegmatis_MC2_155 TPFSKAILTSAYASRGLKMRVTSGGGAEVLMGAAEKCSILYLESRCVSLA
**********************************:***************
Mflv_1245|M.gilvum_PYR-GCK RALGSQGVQNGGIDGVGVVASVPEGMKELLAENLMVMMRDLESCAGNDNL
Mvan_5563|M.vanbaalenii_PYR-1 RALGSQGVQNGGIDGVGVVASVPEGMKELLAENLMVMMRDLESCAGNDNL
MSMEG_6321|M.smegmatis_MC2_155 RALGSQGVQNGGIDGVGVVASVPDGMKELLAENLMVMMRDLESCAGNDNL
***********************:**************************
Mflv_1245|M.gilvum_PYR-GCK ISESDIRRSAHTLPVLLAGADFIFSGFGSIPRYDNAFALSNFNSDDMDDF
Mvan_5563|M.vanbaalenii_PYR-1 ISESDIRRSAHTLPVLLAGADFVFSGFGSIPRYDNAFALSNFNSDDMDDF
MSMEG_6321|M.smegmatis_MC2_155 ISESDIRRSAHTLPVLLAGADFIFSGFGSIPRYDNAFALSNFNSDDMDDF
**********************:***************************
Mflv_1245|M.gilvum_PYR-GCK LVLQRDWGADGGLRTVSPEHLEAVRRRAAQAVQAVYRDLGLADYDDARVE
Mvan_5563|M.vanbaalenii_PYR-1 LVLQRDWGADGGLRTVSPEHLEAVRRRAAKAVQAVYRDLGLADYEDARVE
MSMEG_6321|M.smegmatis_MC2_155 LVLQRDWGADGGLRTVPADQLAAVRRRAARAVQAVYRDLGLADFDDQHIE
****************..::* *******:*************::* ::*
Mflv_1245|M.gilvum_PYR-GCK DVVAANGSRDLPAGHPKMVAEAAASIEARQLTVFDVIAALHRTGFTEEAE
Mvan_5563|M.vanbaalenii_PYR-1 EVVAANGSRDLPAGHPKMVAEAAASIEARQLTVFDVIASLHRTGFTDEAE
MSMEG_6321|M.smegmatis_MC2_155 NVVAANGSRDLPPGDPKAVLEAANAIEAKQLTVFDVVASLKRTGFDPEAE
:***********.*.** * *** :***:*******:*:*:**** ***
Mflv_1245|M.gilvum_PYR-GCK AITTLTRERLRGDQLQTSAIFDEQFRVFSKLTDPNDYRGPATGYALTEQR
Mvan_5563|M.vanbaalenii_PYR-1 AITTLTRERLRGDQLQTSAIFDEKFRVLSKLTDPNDYTGPATGYALTDRR
MSMEG_6321|M.smegmatis_MC2_155 AIMRLTAERMRGDQLQTSAIFDEQFRVLSKITDPNDYAGPGTGYTLSEQR
** ** **:*************:***:**:****** **.***:*:::*
Mflv_1245|M.gilvum_PYR-GCK RAEIDAIRQARSGAELTVDQEEHRGHVVVTDVEPAHQGSDPREVCIGLSP
Mvan_5563|M.vanbaalenii_PYR-1 RAEIDAIRQARSSAELTADQESYRGHVLVTDVEPAQQGSDPREVCIGLSP
MSMEG_6321|M.smegmatis_MC2_155 RAEIDNIRQQRSAAELTADQAEHAGHITVTEIEPARQGSDPREVCIGLSP
***** *** **.****.** .: **: **::***:**************
Mflv_1245|M.gilvum_PYR-GCK AWGRSVWLTLCGLTVGEVLRQIAAGLEEEGCIARTVRVCSTIDTGLIGLT
Mvan_5563|M.vanbaalenii_PYR-1 AWGRSVWLTLCGLTIGEVLRQISAGLEEEGCIARPVRVRSTIDVGLIGLT
MSMEG_6321|M.smegmatis_MC2_155 ALGRSVWLSLCGLPIGEVIRQISAGLEEEGCVPRFVRVRSTIDVGLIGLT
* ******:****.:***:***:********:.* *** ****.******
Mflv_1245|M.gilvum_PYR-GCK AARLSGSGIGIGLQGKGTALIHRRDLAPLANLELFSVAPLLTAKMYRELG
Mvan_5563|M.vanbaalenii_PYR-1 AARLSGSGIGIGLQGKGTALIHRRDLAPLANLELFSVAPLLTAKMYRELG
MSMEG_6321|M.smegmatis_MC2_155 AAKLAGSGIGIGLQGKGTALIHRRDLAPLANLELFSVAPLLTARNYRELG
**:*:**************************************: *****
Mflv_1245|M.gilvum_PYR-GCK KNAARHAKGMAPVPIFTGGTDESISARYHARAVALVALERDACEPGRAPV
Mvan_5563|M.vanbaalenii_PYR-1 KNAARHAKGMAPVPIFTGGTDESISARYHARAVALVALERESCEPGQPPV
MSMEG_6321|M.smegmatis_MC2_155 RNAARHAKGMAPVPILTGGTDESISARYHARAVALVALERQASEPGEAPV
:**************:************************::.***..**
Mflv_1245|M.gilvum_PYR-GCK TVKVEAR
Mvan_5563|M.vanbaalenii_PYR-1 TVKVEWP
MSMEG_6321|M.smegmatis_MC2_155 TVEVRRP
**:*.