For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSQTIFTHDTEITLRAAAELVNSDRVDGEQLADMQSLDAYLDHFGWTGRRDRDDAELQSVQRLRERLGAI WTAADDEEEAVRLVNALLSETKASPWLTRHPEMPDWHLHLASVDDPLWQRMGAEIAMAIADVIRGGELRR LKICAAPDCEAVLTDLSKNRSKRFCDTGNCGNRQHVAAYRQRLRGE
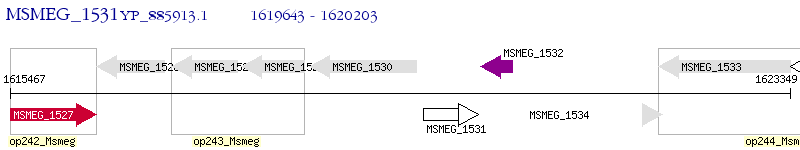
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1531 | - | - | 100% (186) | hypothetical protein MSMEG_1531 |
| M. smegmatis MC2 155 | MSMEG_3408 | - | 2e-10 | 31.78% (129) | hypothetical protein MSMEG_3408 |
| M. smegmatis MC2 155 | MSMEG_0118 | - | 3e-07 | 27.81% (169) | hypothetical protein MSMEG_0118 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4963 | - | 5e-84 | 78.65% (178) | hypothetical protein Mflv_4963 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3761c | - | 1e-65 | 67.05% (173) | hypothetical protein MAB_3761c |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0036 | - | 1e-59 | 67.08% (161) | conserved hypothetical protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_1443 | - | 7e-83 | 78.09% (178) | hypothetical protein Mvan_1443 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4963|M.gilvum_PYR-GCK ---MLFTYDTELTLRAASVLINTDRVDGEQLADLAALDEYLDHFGWTGRR
Mvan_1443|M.vanbaalenii_PYR-1 ---MLFTYDTELTLRAAGVLINSDRVDGDQLSDLAALDEYLDHFGWTGRR
MSMEG_1531|M.smegmatis_MC2_155 MSQTIFTHDTEITLRAAAELVNSDRVDGEQLADMQSLDAYLDHFGWTGRR
MAB_3761c|M.abscessus_ATCC_199 -------------MRSAAELINTGRNDRELLPDVAALPPLLDRYGWTGRR
TH_0036|M.thermoresistible__bu -------------------LVNSDRVAGEQLGAPRDLEAFLDEYGWTGRR
*:*:.* : * * **.:******
Mflv_4963|M.gilvum_PYR-GCK DHDAAELKSVHQLRDRLGRIWAAADDEERAVGHVNALLSDTRASPWLTRH
Mvan_1443|M.vanbaalenii_PYR-1 DHDAAELESVRRLRTRLARIWEAADDETRAVGHVNALLADTKASPWLTRH
MSMEG_1531|M.smegmatis_MC2_155 DRDDAELQSVQRLRERLGAIWTAADDEEEAVRLVNALLSETKASPWLTRH
MAB_3761c|M.abscessus_ATCC_199 DGDDAELEAVKALRARLGDIWARINDEEYVVRQVNALLADTHASPWLTRH
TH_0036|M.thermoresistible__bu DRDAAELRSVHRLRRRLGAIWAVADDEEKTVELVNALLRDTRATPWLTRH
* * ***.:*: ** **. ** :** .* ***** :*:*:******
Mflv_4963|M.gilvum_PYR-GCK PEMP---------------EWHLHLASIHDPLWQRMGAEMAMALADLIRA
Mvan_1443|M.vanbaalenii_PYR-1 PEMP---------------EWHLHLASIHDPLWQRMGAEMAMALADLIRA
MSMEG_1531|M.smegmatis_MC2_155 PEMP---------------DWHLHLASVDDPLWQRMGAEIAMAIADVIRG
MAB_3761c|M.abscessus_ATCC_199 VEEP---------------DWHLHMAASDAPLADRLGAETAMVFADLVRT
TH_0036|M.thermoresistible__bu PEFASSTVAGQRNGLDRQTEWHLHLTAADAPLWQRMGAEIAMALADLIRA
* . :****::: . ** :*:*** **.:**::*
Mflv_4963|M.gilvum_PYR-GCK GELRRLKLCAAPDCEASLIDLSRNRSGKFCDTGNCGNRQHVAAYRERRSN
Mvan_1443|M.vanbaalenii_PYR-1 GELRRLKICAAPDCHAVLVDLSRNRSGKFCDTGNCGNRQHVAAYRERRSN
MSMEG_1531|M.smegmatis_MC2_155 GELRRLKICAAPDCEAVLTDLSKNRSKRFCDTGNCGNRQHVAAYRQRLRG
MAB_3761c|M.abscessus_ATCC_199 GEQRRLKTCAAPDCDAVLVDLSRNRSRMFCDTGNCGNRQHVAAYRERLRD
TH_0036|M.thermoresistible__bu GELRRLKVCAAPDCNAVLIDLSRNRSRRFCXX------------XLRTPG
** **** ******.* * ***:*** ** * .
Mflv_4963|M.gilvum_PYR-GCK K-
Mvan_1443|M.vanbaalenii_PYR-1 K-
MSMEG_1531|M.smegmatis_MC2_155 E-
MAB_3761c|M.abscessus_ATCC_199 EA
TH_0036|M.thermoresistible__bu S-
.