For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MDVAVVDRRDEEFLLDLLNTTPVVDGQERDDLADDDSARAWMRARGVESSRGDLIAVRDLLQDVVRGMRT PSALDRFLGDVTMRPKMTPDGVSWELAASPAARAVLAWDALRINSPGRLRACANDECRLFLIDRSKPNTA RWCSMAICGNRMKARRYYQRAKGSEK
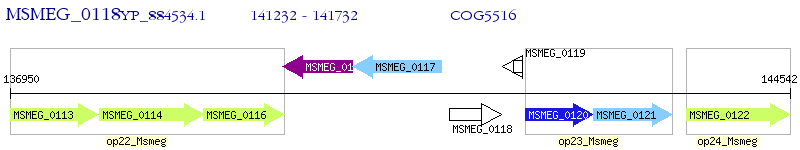
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0118 | - | - | 100% (166) | hypothetical protein MSMEG_0118 |
| M. smegmatis MC2 155 | MSMEG_3613 | - | 7e-13 | 30.23% (172) | hypothetical protein MSMEG_3613 |
| M. smegmatis MC2 155 | MSMEG_3408 | - | 3e-10 | 35.35% (99) | hypothetical protein MSMEG_3408 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4849 | - | 8e-53 | 63.47% (167) | hypothetical protein Mflv_4849 |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3057 | - | 6e-15 | 33.33% (162) | hypothetical protein MAB_3057 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3071 | - | 4e-11 | 30.86% (175) | hypothetical protein Mvan_3071 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0118|M.smegmatis_MC2_155 -MDVAVVDRRDEEFLLDLLNTTPVVDGQER----DDLADDDSARAWMRAR
Mflv_4849|M.gilvum_PYR-GCK -MDVGVASADDEAFLLDLLNTTPVVEGVPT----DVLADKATAAEWMSSH
MAB_3057|M.abscessus_ATCC_1997 --MSDPRPHLGEPLALDLLNTRWMSGDGPR----DLLTDLDGLRVWLHTN
Mvan_3071|M.vanbaalenii_PYR-1 MRANWPDDDEPKPAPQPLRRIQALVNTVERPDGADRLADPSDARPWMIAN
: * . : * *:* *: :.
MSMEG_0118|M.smegmatis_MC2_155 GV--------ESSRGDLIAVRDLLQDVVRGMR--------TPSALDRFLG
Mflv_4849|M.gilvum_PYR-GCK GVG-----ATDKARRVLIDARSALQAVVRGKA--------TPDVLQRFLE
MAB_3057|M.abscessus_ATCC_1997 NLHT-LCAADTQTREALLAAREAIYRAVKDSQ--------IDGLNAVLSH
Mvan_3071|M.vanbaalenii_PYR-1 GLLAPSAELTAADLALAIEVREALRALLIHNAGGPRPGPEAVAPLQRIAA
.: : .*. : : :
MSMEG_0118|M.smegmatis_MC2_155 DVTMRPKMTPDGVSWELAASP-AARAVLAWD-----ALRINSPGRLRACA
Mflv_4849|M.gilvum_PYR-GCK GVHLTPTATDGGVDWTLAGTDGAARAVLAWD-----GLRITSPGRLRPCA
MAB_3057|M.abscessus_ATCC_1997 GAIRRSLTANGPLDTAEVTESHWLPGWLAADNLL--ALLSESPDRIKQCA
Mvan_3071|M.vanbaalenii_PYR-1 KGRLQAAVGDGGVVTLRVDGDSVADRLAGLLPVIRDGQRDGTWALLKACG
. . : . . . : :: *.
MSMEG_0118|M.smegmatis_MC2_155 NDECRLFLIDRSKPNTARWCSMAICGNRMKARRYYQRAKGSEK-------
Mflv_4849|M.gilvum_PYR-GCK NTECQLFLIDRSKPNNARWCSMAICGNRMKARRHYQRSRG----------
MAB_3057|M.abscessus_ATCC_1997 HPQCVLFFYDTSKNGARRWHSMTTCGNRAKAARHYAKNT-----------
Mvan_3071|M.vanbaalenii_PYR-1 NDECRWVFYDSSRNHGGTWCDMASCGNKLKNRQFRARRRADGPSGQVPHQ
: :* .: * *: * .*: ***: * :. :
MSMEG_0118|M.smegmatis_MC2_155 ----------
Mflv_4849|M.gilvum_PYR-GCK ----------
MAB_3057|M.abscessus_ATCC_1997 ----------
Mvan_3071|M.vanbaalenii_PYR-1 GGGQRAQPVE