For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VINLRGRRKPKVVTQRTSAELDAMAVAGALVASALRAVKAAAAPGVSTLELDRIAETVIRDGGGTPSFLG YHGFPASICASVNERVVHGIPSADEVLAAGDLVSIDCGAIVDGWHGDSAVTFGVGPLIPADEALSEATKA AMEAGIAAMVPGNRLTDVSHAIEVETHAAEKRDDRKYGIVANYGGHGIGRQMYMDPFLPNEGAPGRGPYL APGSVLAIEPMLTLGTTKTVVLDDEWTVVTADGTRAAHWEHTVAVTEDGPRILTQ
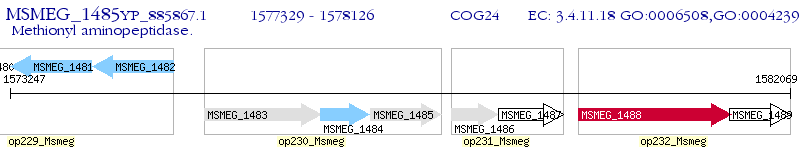
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1485 | map | - | 100% (265) | methionine aminopeptidase |
| M. smegmatis MC2 155 | MSMEG_5683 | map | 7e-63 | 52.31% (260) | methionine aminopeptidase, type I |
| M. smegmatis MC2 155 | MSMEG_5050 | map | 5e-50 | 42.47% (259) | methionine aminopeptidase, type I |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0755 | mapA | 1e-117 | 78.54% (261) | methionine aminopeptidase |
| M. gilvum PYR-GCK | Mflv_5020 | - | 1e-125 | 82.20% (264) | methionine aminopeptidase |
| M. tuberculosis H37Rv | Rv0734 | mapA | 1e-117 | 78.54% (261) | methionine aminopeptidase |
| M. leprae Br4923 | MLBr_01831 | mapA | 1e-110 | 73.54% (257) | methionine aminopeptidase |
| M. abscessus ATCC 19977 | MAB_3782c | - | 1e-111 | 75.29% (263) | methionine aminopeptidase |
| M. marinum M | MMAR_1072 | mapA | 1e-116 | 77.78% (261) | methionine aminopeptidase, MapA |
| M. avium 104 | MAV_4432 | map | 1e-116 | 77.78% (261) | methionine aminopeptidase |
| M. thermoresistible (build 8) | TH_3403 | - | 1e-126 | 84.09% (264) | PUTATIVE methionine aminopeptidase, type I |
| M. ulcerans Agy99 | MUL_0830 | mapA | 1e-116 | 77.39% (261) | methionine aminopeptidase |
| M. vanbaalenii PYR-1 | Mvan_1349 | - | 1e-124 | 81.44% (264) | methionine aminopeptidase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1485|M.smegmatis_MC2_155 MINLRGRRKPKVVTQRTSAELDAMAVAGALVASALRAVKAAAAPGVSTLE
TH_3403|M.thermoresistible__bu LVGLRLRNR-KVVAQRSAGELDAMAAAGALVAEALRAVQAAAAPGVSTKH
Mflv_5020|M.gilvum_PYR-GCK MVSLPGLRSRKVVPQRTAGELDAMAAAGALVAAALKAVQAEAAPGKSTLD
Mvan_1349|M.vanbaalenii_PYR-1 MVSLPGLRSRKVVPQRSAGELDAMAAAGALVAAALKAVRAAAAPGVSTLE
MMAR_1072|M.marinum_M MRALARLRGRKVVPQRTAGELDAMAAAGAVVAAAHQAVRSAAVAGVSTLQ
MUL_0830|M.ulcerans_Agy99 MRALARLRGSRVVPQRTAGELDAMAAAGAVVAAAHQAVRSAAVAGVSTLQ
Mb0755|M.bovis_AF2122/97 MRPLARLRGRRVVPQRSAGELDAMAAAGAVVAAALRAIRAAAAPGTSSLS
Rv0734|M.tuberculosis_H37Rv MRPLARLRGRRVVPQRSAGELDAMAAAGAVVAAALRAIRAAAAPGTSSLS
MLBr_01831|M.leprae_Br4923 MGALTRLRSRRVVPQRSVGELDAMAAAGAVVAAALHAVRDAAGPGVSSLS
MAV_4432|M.avium_104 MNALARLRGRKVVPQRSDGELDAMAAAGAVVAAALQAVRAAAVAGVSTLS
MAB_3782c|M.abscessus_ATCC_199 ---MGLFGRKKTVEQRTPGELDAMAAAGSIVGAALVAVRDAAKAGVSTLE
: :.* **: .******.**::*. * *:: * .* *:
MSMEG_1485|M.smegmatis_MC2_155 LDRIAETVIRDGGGTPSFLGYHGFPASICASVNERVVHGIPSADEVLAAG
TH_3403|M.thermoresistible__bu LDEIAESVIRDGGGIPSFLGYHGFPASICASVNERVVHGIPADDEILAAG
Mflv_5020|M.gilvum_PYR-GCK LDRVAEAVIRDGGGVPSFLGYHGFPATICASVNDRVVHGIPTADETLAAG
Mvan_1349|M.vanbaalenii_PYR-1 LDEIAESVIRDGGGIPSFLGYHGFPASICASVNDRVVHGIPTAAEKLAAG
MMAR_1072|M.marinum_M LDEIAESVIREAGATPSFLGYHGFPASICASVNDRVVHGIPSATEILGPG
MUL_0830|M.ulcerans_Agy99 LDEIAESVIREAGATPSFLGYHGFPASICASVNDRVVHGIPSATEILGPG
Mb0755|M.bovis_AF2122/97 LDEIAESVIRESGATPSFLGYHGYPASICASINDRVVHGIPSTAEVLAPG
Rv0734|M.tuberculosis_H37Rv LDEIAESVIRESGATPSFLGYHGYPASICASINDRVVHGIPSTAEVLAPG
MLBr_01831|M.leprae_Br4923 LDEIAESVIREAGATPSFLRYHGYPATICTSVNDRVIHGIPSAVEILAPG
MAV_4432|M.avium_104 LDQIAESVIRDAGAVPSFLGYHGYPASICASVNDRVVHGIPSGAEVLAPG
MAB_3782c|M.abscessus_ATCC_199 LDQVAESVIREAGAVPSFLGYHGFPASICSSVNDQVVHGIPSATAVLADG
**.:**:***:.*. **** ***:**:**:*:*::*:****: *. *
MSMEG_1485|M.smegmatis_MC2_155 DLVSIDCGAIVDGWHGDSAVTFGVGPLIPADEALSEATKAAMEAGIAAMV
TH_3403|M.thermoresistible__bu DLVSIDCGAILDGWHGDSAITFGVGDLIPADEALSQATKAALDAGIDAMR
Mflv_5020|M.gilvum_PYR-GCK DLVSIDCGAILDGWHGDSALTFGVGDLIPADEALSAATRESMEAGIAAMV
Mvan_1349|M.vanbaalenii_PYR-1 DLVSIDCGAILDGWHGDSAVTFGVGEVIPVDESLSAATRESMEAGIAAMV
MMAR_1072|M.marinum_M DLVSIDCGAVLDGWHGDAAITFGIGTLIPVDEALSEATRESLEAGIAAMV
MUL_0830|M.ulcerans_Agy99 DLVSIDCGAVLDGWHGDAAITFGIGTLIPVDEALSEATRESLEAGIAAMV
Mb0755|M.bovis_AF2122/97 DLVSIDCGAVLDGWHGDAAITFGVGALSDADEALSEATRESLQAGIAAMV
Rv0734|M.tuberculosis_H37Rv DLVSIDCGAVLDGWHGDAAITFGVGALSDADEALSEATRESLQAGIAAMV
MLBr_01831|M.leprae_Br4923 DLVSIDCGAVLDGWHGDAAITFGVGTLTSVDEALSQATRESLEAGIAAMI
MAV_4432|M.avium_104 DLVSMDCGAVLDGWHGDAAITFGVGALDPADEALRQATRESLEAGIAAMV
MAB_3782c|M.abscessus_ATCC_199 DLVSIDCGAILDGWHGDSAWTFAVGTVIPSDEALSEATRLSMEAGIAAMI
****:****::******:* **.:* : **:* **: :::*** **
MSMEG_1485|M.smegmatis_MC2_155 PGNRLTDVSHAIEVETHAAEKRDDRKYGIVANYGGHGIGRQMYMDPFLPN
TH_3403|M.thermoresistible__bu PGNRLTDVSHAIETATRAAERRDNRRYGIVANYGGHGIGRQMHMDPFLPN
Mflv_5020|M.gilvum_PYR-GCK PGNRLTDVSHAIEQGTRAAEARHGRKFGIVAGYGGHGIGREMHMDPFLAN
Mvan_1349|M.vanbaalenii_PYR-1 PGNRLSDVSHAIEQGTRAAAQRHGRKFGIVAGYGGHGIGREMHMDPFLPN
MMAR_1072|M.marinum_M IGNRLTDVAHAIETETRAAEARYQRSFGIVDGYGGHGIGREMHMDPFLPN
MUL_0830|M.ulcerans_Agy99 IGNRLTDVAHAIETETRAAEARYQRSFGIVDGYGGHGIGREMHMDPFLPN
Mb0755|M.bovis_AF2122/97 VGNRLTDVAHAIETGTRAAELRYGRSFGIVAGYGGHGIGRQMHMDPFLPN
Rv0734|M.tuberculosis_H37Rv VGNRLTDVAHAIETGTRAAELRYGRSFGIVAGYGGHGIGRQMHMDPFLPN
MLBr_01831|M.leprae_Br4923 VNNRLTDVAHAIEVSTRTSEIRYRLTFGIVQGYGGHGIGRHMHMDPFLPN
MAV_4432|M.avium_104 PGNRLTDVSHAIELGTRAAETRHGRAFGIVEGYGGHGIGRQMHMDPFLPN
MAB_3782c|M.abscessus_ATCC_199 PGNRLTDVSHAIELGTRAAEKQFDRAFGIVDGYGGHGIGRSMHLDPFLPN
.***:**:**** *::: : :*** .******** *::****.*
MSMEG_1485|M.smegmatis_MC2_155 EGAPGRGPYLAPGSVLAIEPMLTLGTTKTVVLDDEWTVVTADGTRAAHWE
TH_3403|M.thermoresistible__bu EGAPGRGPHLAVGSVLAIEPMLTLGTTETVVLADEWTVVTADGSRAAHWE
Mflv_5020|M.gilvum_PYR-GCK EGAPGRGPYLAPGSVLAIEPMLTLGTRKTLILDDGWTVVTADGTRAAHWE
Mvan_1349|M.vanbaalenii_PYR-1 EGSPGRGPYLAPGSVLAIEPMLTLGTAKTIVLDDGWTVVTADGSRAAHWE
MMAR_1072|M.marinum_M EGAPGRGPTLVAGSVLAIEPMLTLGTRKTVVLDDQWTVITADGSRAAHWE
MUL_0830|M.ulcerans_Agy99 EGAPGRGPTLVAGSVLAIEPMLTLGTRKTVVLDDQWTVITADGSRAAHWE
Mb0755|M.bovis_AF2122/97 EGAPGRGPLLAAGSVLAIEPMLTLGTTKTVVLDDKWTVTTADGSRAAHWE
Rv0734|M.tuberculosis_H37Rv EGAPGRGPLLAAGSVLAIEPMLTLGTTKTVVLDDKWTVTTADGSRAAHWE
MLBr_01831|M.leprae_Br4923 EGAPGRGPLLVPGSVLAIEPMLTLGTGKTIVLDDEWTVTTADGSRAAHWE
MAV_4432|M.avium_104 EGSPGRGPLLAPGSVLAIEPMLTLGTGKTVVLDDQWTVITTDGSRAAHWE
MAB_3782c|M.abscessus_ATCC_199 EGAPGKGPLLAVGSVLAIEPMLTLGTTQTRVLADDWTVVTTDGSRAAHWE
**:**:** *. ************** :* :* * *** *:**:******
MSMEG_1485|M.smegmatis_MC2_155 HTVAVTEDGPRILTQ--
TH_3403|M.thermoresistible__bu HTVAVTEDGPRILTL--
Mflv_5020|M.gilvum_PYR-GCK HTVAVTDDGPRILTAR-
Mvan_1349|M.vanbaalenii_PYR-1 HTVAVTDDGPRILTV--
MMAR_1072|M.marinum_M HTVAVTENGPRILTLG-
MUL_0830|M.ulcerans_Agy99 HTVAVTENGPRILTLG-
Mb0755|M.bovis_AF2122/97 HTVAVTDDGPRILTLG-
Rv0734|M.tuberculosis_H37Rv HTVAVTDDGPRILTLG-
MLBr_01831|M.leprae_Br4923 HTVAVTEDGPRVLTLA-
MAV_4432|M.avium_104 HTVAVTDAGPRILTLG-
MAB_3782c|M.abscessus_ATCC_199 HTVAVTEAGPRILTMRP
******: ***:**