For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLTELVEVPGGSFRMGSTSFYPEEAPVHTATVGDFAIERHPVTNAQFAEFVEATGYVTVAERPLDPKLYP GVPEADLVPGSLVFRPTSGPVDLRDWRQWWDWAPGACWHHPFGPRREFCRPDHPVVQVAYPDAVAYATWA GRRLPTEAEWEYAARGGPKGGVGFLYAWGDEVCPDGQLMANTWQGKFPYRNDGALGWKGTSPVGTFPPNR LGLVDMIGNVWEWTATKFSAHHRPGDESHVTCCPPPSGGDPGVNQVLKGGSHLCAPEYCHRYRPAARSPQ SQDSSTTHIGFRCVVS
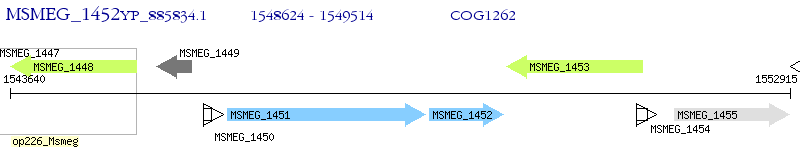
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1452 | - | - | 100% (296) | sulfatase-modifying factor 1 |
| M. smegmatis MC2 155 | MSMEG_1226 | - | e-134 | 74.58% (295) | sulfatase-modifying factor 1 |
| M. smegmatis MC2 155 | MSMEG_6249 | - | 8e-20 | 30.80% (276) | hypothetical protein MSMEG_6249 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0733 | - | 1e-135 | 74.50% (298) | hypothetical protein Mb0733 |
| M. gilvum PYR-GCK | Mflv_5038 | - | 1e-134 | 74.41% (297) | hypothetical protein Mflv_5038 |
| M. tuberculosis H37Rv | Rv0712 | - | 1e-135 | 74.50% (298) | hypothetical protein Rv0712 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4305 | - | 1e-127 | 71.77% (294) | hypothetical protein MAB_4305 |
| M. marinum M | MMAR_1043 | - | 1e-137 | 76.61% (295) | hypothetical protein MMAR_1043 |
| M. avium 104 | MAV_4458 | - | 1e-136 | 75.43% (293) | sulfatase-modifying factor 1 |
| M. thermoresistible (build 8) | TH_0314 | - | 1e-135 | 74.25% (299) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_0225 | - | 3e-85 | 53.74% (294) | hypothetical protein MUL_0225 |
| M. vanbaalenii PYR-1 | Mvan_1319 | - | 1e-129 | 72.64% (296) | hypothetical protein Mvan_1319 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_5038|M.gilvum_PYR-GCK ------------------MLTDLVDLEGGSFRMGSTRFYPEEAPAHTVTV
Mvan_1319|M.vanbaalenii_PYR-1 ------------------MLTELVALDGGSFRMGSTRFYPEEAPVHTVTV
TH_0314|M.thermoresistible__bu ----------------MSALTDLVEIPGGTFRMGSIRFYPEEAPVHTVTV
MSMEG_1452|M.smegmatis_MC2_155 ------------------MLTELVEVPGGSFRMGSTSFYPEEAPVHTATV
Mb0733|M.bovis_AF2122/97 ------------------MLTELVDLPGGSFRMGSTRFYPEEAPIHTVTV
Rv0712|M.tuberculosis_H37Rv ------------------MLTELVDLPGGSFRMGSTRFYPEEAPIHTVTV
MMAR_1043|M.marinum_M ------------------MLTELVDLPGGSFRMGSTSFYPEEAPIHSVSV
MAV_4458|M.avium_104 ------------------MRSELADLPGGSFRMGSTSFYPEEAPIHTVTV
MAB_4305|M.abscessus_ATCC_1997 MEYSPRNRKLREMETAAVTLTELVELPGGSFPMGCNVFYPEEMPERVSSV
MUL_0225|M.ulcerans_Agy99 ------------------MTDDLIRIPAQTAVLGSEAHYPEESPLRQVDV
:* : . : :*. .**** * : *
Mflv_5038|M.gilvum_PYR-GCK APFAIERNPVTNAQFAEFVDDTGYRTVAERPPDPALYPGASPHDLVPGAL
Mvan_1319|M.vanbaalenii_PYR-1 APYAIERHPVTNAQFASFVADTGYRTVAELAPDPALYPGAAPQDLVPGAL
TH_0314|M.thermoresistible__bu ADFAIERHPVTNAQFAEFVAATGYRTVAEQPLDPRLYPGVAAADLQPGAL
MSMEG_1452|M.smegmatis_MC2_155 GDFAIERHPVTNAQFAEFVEATGYVTVAERPLDPKLYPGVPEADLVPGSL
Mb0733|M.bovis_AF2122/97 RAFAVERHPVTNAQFAEFVSATGYVTVAEQPLDPGLYPGVDAADLCPGAM
Rv0712|M.tuberculosis_H37Rv RAFAVERHPVTNAQFAEFVSATGYVTVAEQPLDPGLYPGVDAADLCPGAM
MMAR_1043|M.marinum_M HAFAIERYPVTNAQFAEFVAATGYVTVAEQPLDPALYPGVNPDDLRPGAM
MAV_4458|M.avium_104 EPFAIERHPVTNAQFAEFVAATGYVTVAEQPIDPALYPGADPSQLQPGAM
MAB_4305|M.abscessus_ATCC_1997 EPFAIERHPVTNAQFAEFVTQTGYVTVAERAPDPDMYPGASPDDLVPGAM
MUL_0225|M.ulcerans_Agy99 NAFRIQAHQVTNTQFAEFVDATGYLTVAERPLDPADYPGAPPENLQPGSM
: :: ***:***.** *** **** . ** ***. :* **::
Mflv_5038|M.gilvum_PYR-GCK VFRPTAGPVDLRDWRQWWEWTPGADWRHPFGP-DSSVDDRPEHPVVQVCY
Mvan_1319|M.vanbaalenii_PYR-1 VFRPTAGPVDLTDWRQWWEWVPGADWRHPSGP-GSSVDDKPDHPVVQVCY
TH_0314|M.thermoresistible__bu VFTPTRGPVDLRDWRQWWTWTPGACWRHPFGPEGPSWQDRPDHPVVQIAY
MSMEG_1452|M.smegmatis_MC2_155 VFRPTSGPVDLRDWRQWWDWAPGACWHHPFGP--RREFCRPDHPVVQVAY
Mb0733|M.bovis_AF2122/97 VFCPTAGPVDLRDWRQWWDWVPGACWRHPFGR-DSDIADRAGHPVVQVAY
Rv0712|M.tuberculosis_H37Rv VFCPTAGPVDLRDWRQWWDWVPGACWRHPFGR-DSDIADRAGHPVVQVAY
MMAR_1043|M.marinum_M VFRPTAGPVDLRDWRQWWDWAPGASWRHPWGP-DSDVDDRADHPVVQVAY
MAV_4458|M.avium_104 VFRPTPGPVDLRDWRQWWHWVPGASWRHPFGP-DSDVADRADHPVVQVAY
MAB_4305|M.abscessus_ATCC_1997 TFRTTAGPVDLHDWQQWWDWVPGAYWRHPFGP-GSDIDGAAEHPVVQVAY
MUL_0225|M.ulcerans_Agy99 VFTRTAGPVDLRHLSLWWTWTPGASWRHPAGP-HSSIAKRADHPVVHVAY
.* * ***** . ** *.*** *:** * . ****::.*
Mflv_5038|M.gilvum_PYR-GCK SDAAAYARWAGRRLPTEAEWEFAAGAGSTA----VYAWGDEPAPGGQLMA
Mvan_1319|M.vanbaalenii_PYR-1 ADAAAYARWAGRRLPTEAEWEFAARAGSTA----VYAWGDEAVPGGALMA
TH_0314|M.thermoresistible__bu PDAQAYARWAGRRLPTEREWEYAARAGADT----VYPWGDEARPGGRLMA
MSMEG_1452|M.smegmatis_MC2_155 PDAVAYATWAGRRLPTEAEWEYAARGGPKGGVGFLYAWGDEVCPDGQLMA
Mb0733|M.bovis_AF2122/97 PDAVAYARWAGRRLPTEAEWEYAARGGTTA----TYAWGDQEKPGGMLMA
Rv0712|M.tuberculosis_H37Rv PDAVAYARWAGRRLPTEAEWEYAARGGTTA----TYAWGDQEKPGGMLMA
MMAR_1043|M.marinum_M PDAAAYARWAGRRLPTEAEWEYAARGATTT----TYAWGDEEKPAGQLMA
MAV_4458|M.avium_104 PDAAAYARWAGRRLPTEAEWEYAARAGATT----TYPWGDEPTSDGRLMA
MAB_4305|M.abscessus_ATCC_1997 TDAAAYARWAGRRLPTEAEWEYAARGGSGT----VYAWGDEVAPEGRLMA
MUL_0225|M.ulcerans_Agy99 EDAHAYAGWAGLALPSEAEWEVAARGGLTG---AAYTWGEESERPGQRLA
** *** *** **:* *** ** .. *.**:: * :*
Mflv_5038|M.gilvum_PYR-GCK NTWQGAFPYRNDGALGWAGTSPVGTFPANGFGLVDMIGNVWEWTTTRFAG
Mvan_1319|M.vanbaalenii_PYR-1 NTWQGAFPYRNDGALGWSGTSPVGTFPANAYGLVDMIGNVWEWTTTRFAG
TH_0314|M.thermoresistible__bu NTWQGRFPYRNDGAAGWIGTSPVGAFPPNDFGLFDMIGNVWEWTTTRFSG
MSMEG_1452|M.smegmatis_MC2_155 NTWQGKFPYRNDGALGWKGTSPVGTFPPNRLGLVDMIGNVWEWTATKFSA
Mb0733|M.bovis_AF2122/97 NTWQGRFPYRNDGALGWVGTSPVGRFPANGFGLLDMIGNVWEWTTTEFYP
Rv0712|M.tuberculosis_H37Rv NTWQGRFPYRNDGALGWVGTSPVGRFPANGFGLLDMIGNVWEWTTTEFYP
MMAR_1043|M.marinum_M NTWQGKFPYRNDGALGWFGTSPVGTFPANGFGLFDMIGNVWEWTTTPFSA
MAV_4458|M.avium_104 NTWQGRFPYRNDGALGWVGTSPVGVFAPNAFGLLDMIGNVWEWTTTEFST
MAB_4305|M.abscessus_ATCC_1997 NTWQGRFPYRNDGALGWHGTSPVGTFPANDFGLLDMIGNVWEWTSTRFQ-
MUL_0225|M.ulcerans_Agy99 NFWHGDFPWRPE--PGYGRTSPVGSFEPNGYGLFDMAGNVWEWTVDWYID
* *:* **:* : *: ***** * .* **.** ******* :
Mflv_5038|M.gilvum_PYR-GCK HHR---VNPPPPPTCCPPAD----PDPAVNQALKGGSHLCAPEYCHRYRP
Mvan_1319|M.vanbaalenii_PYR-1 HHV---LDAPVN-SCCPQAG----PDPAVTQTLKGGSHLCAPEYCHRYRP
TH_0314|M.thermoresistible__bu HHRPAEADEPVR-SCCTPSAPVERPDPAVSQTLKGGSHLCAPEYCHRYRA
MSMEG_1452|M.smegmatis_MC2_155 HHR---PGDESHVTCCPPPS---GGDPGVNQVLKGGSHLCAPEYCHRYRP
Mb0733|M.bovis_AF2122/97 HHR---IDPPST-ACCAPVKLATAADPTISQTLKGGSHLCAPEYCHRYRP
Rv0712|M.tuberculosis_H37Rv HHR---IDPPST-ACCAPVKLATAADPTISQTLKGGSHLCAPEYCHRYRP
MMAR_1043|M.marinum_M HHR---IDQPPK-ACCAP---AGPADPTISQTLKGGSHLCAPEYCHRYRP
MAV_4458|M.avium_104 HHR---IGAATK-PCCAP---SGPADPGVNQTLKGGSHLCAPEYCHRYRP
MAB_4305|M.abscessus_ATCC_1997 -HG---AGAPKS--CCSPSD---NPDPSVIQVLKGGSHLCAPEYCHRYRP
MUL_0225|M.ulcerans_Agy99 TPS---ARPAAEADSYDPQQ---PQFRVPRKVVKGGSFLCADSYCRRYRP
. :.:****.*** .**:***.
Mflv_5038|M.gilvum_PYR-GCK SARSPQSQDSATTHIGFRCAASPV--
Mvan_1319|M.vanbaalenii_PYR-1 SARSPQSQDSATTHIGFRCAV-----
TH_0314|M.thermoresistible__bu SARSPQSQDSAATHIGFRCAA-----
MSMEG_1452|M.smegmatis_MC2_155 AARSPQSQDSSTTHIGFRCVVS----
Mb0733|M.bovis_AF2122/97 AARSPQSQDTATTHIGFRCVADPVSG
Rv0712|M.tuberculosis_H37Rv AARSPQSQDTATTHIGFRCVADPVSG
MMAR_1043|M.marinum_M AARSPQSQDTATTHIGFRCVADR---
MAV_4458|M.avium_104 AARSPQSQDSATTHIGFRCVAEVGSN
MAB_4305|M.abscessus_ATCC_1997 AARSPQSQDSATTHIGFRCVENLG--
MUL_0225|M.ulcerans_Agy99 AARRPQPIDTGVSHIGFRCVGRD---
:** **. *:..:******.