For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VGQKINPHGFRLGITTEWKSRWYADKQYKDYVKEDVAIRKLLATGLERAGIADVEIERTRDRVRVDIHTA RPGIVIGRRGTEADRIRADLEKLTGKQVQLNILEVKNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSA MRQPNVKGIRVQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKVWIYKGDIVGG KRELAAAAPASDRPRRERPSGTRPRRSGSAGTTATSTEAGRAATSDAPAAGTAAAAEAPAESTES
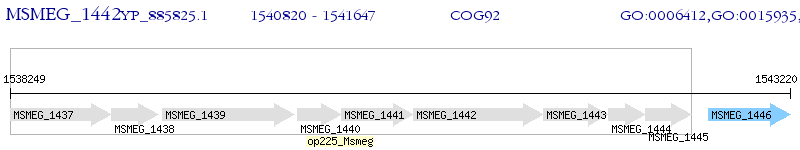
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1442 | rpsC | - | 100% (275) | 30S ribosomal protein S3 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0727 | rpsC | 1e-139 | 90.94% (276) | 30S ribosomal protein S3 |
| M. gilvum PYR-GCK | Mflv_5043 | rpsC | 1e-142 | 92.50% (280) | 30S ribosomal protein S3 |
| M. tuberculosis H37Rv | Rv0707 | rpsC | 1e-139 | 90.94% (276) | 30S ribosomal protein S3 |
| M. leprae Br4923 | MLBr_01857 | rpsC | 1e-133 | 90.80% (261) | 30S ribosomal protein S3 |
| M. abscessus ATCC 19977 | MAB_3814c | rpsC | 1e-137 | 89.64% (280) | 30S ribosomal protein S3 |
| M. marinum M | MMAR_1037 | rpsC | 1e-139 | 90.58% (276) | 30S ribosomal protein S3, RpsC |
| M. avium 104 | MAV_4465 | rpsC | 1e-140 | 90.71% (280) | 30S ribosomal protein S3 |
| M. thermoresistible (build 8) | TH_0401 | rpsC | 1e-131 | 86.52% (282) | PROBABLE 30S RIBOSOMAL PROTEIN S3 RPSC |
| M. ulcerans Agy99 | MUL_0796 | rpsC | 1e-139 | 90.58% (276) | 30S ribosomal protein S3 |
| M. vanbaalenii PYR-1 | Mvan_1311 | rpsC | 1e-141 | 91.13% (282) | 30S ribosomal protein S3 |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1442|M.smegmatis_MC2_155 MGQKINPHGFRLGITTEWKSRWYADKQYKDYVKEDVAIRKLLATGLERAG
MAB_3814c|M.abscessus_ATCC_199 MGQKINPHGFRLGITTDWKSRWYADKQYADYVKEDVAIRRLLATGLERAG
Mflv_5043|M.gilvum_PYR-GCK MGQKINPHGFRLGITTDWKSRWYADKQYADYIKEDVAIRKLLATGLERAG
Mvan_1311|M.vanbaalenii_PYR-1 MGQKINPHGFRLGITTEWKSRWYADKQYADYIKEDVAIRKLLATGLERAG
Mb0727|M.bovis_AF2122/97 MGQKINPHGFRLGITTDWKSRWYADKQYAEYVKEDVAIRRLLSSGLERAG
Rv0707|M.tuberculosis_H37Rv MGQKINPHGFRLGITTDWKSRWYADKQYAEYVKEDVAIRRLLSSGLERAG
MLBr_01857|M.leprae_Br4923 MGQKINPHGFRLGITTGWKSRWYADKQYAEYVKEDVAIRRLLSTGLERAG
MAV_4465|M.avium_104 MGQKINPHGFRLGITTDWKSRWYADKQYADYVKEDVAIRRLLSTGLERAG
MMAR_1037|M.marinum_M MGQKINPHGFRLGITTDWKSRWYADKQYADYVKEDVAIRRLLSSGLERAG
MUL_0796|M.ulcerans_Agy99 MGQKINPHGFRLGITTDWKSRWYADKQYADYVKEDVAIRRLLSSGLERAG
TH_0401|M.thermoresistible__bu VGQKINPHGFRLGITTDWKSRWYADKQYKDYVKEDVAIRRLLSTGLERAG
:*************** *********** :*:*******:**::******
MSMEG_1442|M.smegmatis_MC2_155 IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTGKQVQL
MAB_3814c|M.abscessus_ATCC_199 IAKVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTGKQVQL
Mflv_5043|M.gilvum_PYR-GCK IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTKKQVQL
Mvan_1311|M.vanbaalenii_PYR-1 IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTKKQVQL
Mb0727|M.bovis_AF2122/97 IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTGKQVQL
Rv0707|M.tuberculosis_H37Rv IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTGKQVQL
MLBr_01857|M.leprae_Br4923 IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTCKQVQL
MAV_4465|M.avium_104 IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTGKQVQL
MMAR_1037|M.marinum_M IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTGKQVQL
MUL_0796|M.ulcerans_Agy99 IADVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRADLEKLTGKQVQL
TH_0401|M.thermoresistible__bu IANVEIERTRDRVRVDIHTARPGIVIGRRGTEADRIRGDLEKLTGKQVQL
**.**********************************.****** *****
MSMEG_1442|M.smegmatis_MC2_155 NILEVKNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
MAB_3814c|M.abscessus_ATCC_199 NILEVKNPEAEAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
Mflv_5043|M.gilvum_PYR-GCK NILEVKNPESRAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
Mvan_1311|M.vanbaalenii_PYR-1 NILEVKNPESVAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
Mb0727|M.bovis_AF2122/97 NILEVKNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
Rv0707|M.tuberculosis_H37Rv NILEVKNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
MLBr_01857|M.leprae_Br4923 NILEVKNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
MAV_4465|M.avium_104 NILEVRNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
MMAR_1037|M.marinum_M NILEVKNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
MUL_0796|M.ulcerans_Agy99 NILEVKNPESQAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
TH_0401|M.thermoresistible__bu NILEVKNPESNAQLVAQGVAEQLSNRVAFRRAMRKAIQSAMRQPNVKGIR
*****:***: ***************************************
MSMEG_1442|M.smegmatis_MC2_155 VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
MAB_3814c|M.abscessus_ATCC_199 VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
Mflv_5043|M.gilvum_PYR-GCK VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
Mvan_1311|M.vanbaalenii_PYR-1 VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
Mb0727|M.bovis_AF2122/97 VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
Rv0707|M.tuberculosis_H37Rv VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
MLBr_01857|M.leprae_Br4923 VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLHEAKTTFGRIGVKV
MAV_4465|M.avium_104 VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
MMAR_1037|M.marinum_M VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
MUL_0796|M.ulcerans_Agy99 VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEAKTTFGRIGVKV
TH_0401|M.thermoresistible__bu VQCSGRLGGAEMSRSEFYREGRVPLHTLRADIDYGLYEARTTFGRIGVKV
************************************:**:**********
MSMEG_1442|M.smegmatis_MC2_155 WIYKGDIVGGKRELAAAAPAS-----DRPRRERPSGTRPRRSGSAGTTAT
MAB_3814c|M.abscessus_ATCC_199 WIYKGDIVGGKRELAAAGVEAGRGAPDRPRRERPAGTRPRRSGSSGTTAT
Mflv_5043|M.gilvum_PYR-GCK WIYKGDIVGGKRELAAAAPAG----ADRPRRERPSGTRPRRSGASGTTAT
Mvan_1311|M.vanbaalenii_PYR-1 WIYKGDIVGGKRELTAAAPA-----ADRPRRDRPSGTRPRRSGASGTTAT
Mb0727|M.bovis_AF2122/97 WIYKGDIVGGKRELAAAAPAG----ADRPRRERPSGTRPRRSGASGTTAT
Rv0707|M.tuberculosis_H37Rv WIYKGDIVGGKRELAAAAPAG----ADRPRRERPSGTRPRRSGASGTTAT
MLBr_01857|M.leprae_Br4923 WIYKGDIVGGKREVTAVAPAG----AERARRERPSGTRPRRSGAAGTTVT
MAV_4465|M.avium_104 WIYKGDVVGGKRELAAAAPAG----AERPRRERPSGTRPRRSGASGTTAT
MMAR_1037|M.marinum_M WIYKGDIVGGKRELAAAVPAG----ADRPRRERPAGSRPRRSGASGTTAT
MUL_0796|M.ulcerans_Agy99 WIYKGDIVGGKRELAAAVPAG----ADRPRRERPAGSRPRRSGASGTTAT
TH_0401|M.thermoresistible__bu WIYKGDVVGGKRELGVAPPSE------RPRRERPS--RPRRSGASGTTAT
******:******: .. *.**:**: ******::***.*
MSMEG_1442|M.smegmatis_MC2_155 ------STEAGRAATS----DAPAAGT---AAAAEAP-AESTES---
MAB_3814c|M.abscessus_ATCC_199 ------STEAGRAAAE----TPASDG------ASAPS-AETTES---
Mflv_5043|M.gilvum_PYR-GCK ------STDAGRAATE---EAPATDAA---ATAPAAGQPETTES---
Mvan_1311|M.vanbaalenii_PYR-1 ------STDAGRAASEGTVEAPATEAA---ATAPSAGQPETTES---
Mb0727|M.bovis_AF2122/97 ------GTDAGRAAG---GEEAAP-------DAAAPVEAQSTES---
Rv0707|M.tuberculosis_H37Rv ------GTDAGRAAG---GEEAAP-------DAAAPVEAQSTES---
MLBr_01857|M.leprae_Br4923 ------GTDAGRAVG---GQESAATNIGHSDDSVVTHEPQIAES---
MAV_4465|M.avium_104 ------GTEAGRAAAS--ADESTAAG--QPAEAAPAAEPQSTES---
MMAR_1037|M.marinum_M ------GTEAGRAVGS---EEPAAA------ESATTPEAQSTES---
MUL_0796|M.ulcerans_Agy99 ------GTEAGRAVGS---EEPAAA------ESATTPEAQSTES---
TH_0401|M.thermoresistible__bu GTTPPPGTEQAPATAEASAGTEAAAG-----AAAAAPSTETSTSGES
.*: . *. .. : . .: : *