For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
YAEERQQAIAALVLSRGRASVAELAQAYDVTTETVRRDLAVLDRAGVVRRVHGGAVPVRTLHLVEAGVGE RDSTRAEQKDAIAAAAAEFFPPTGSSVLLDAGTTTARVAAHLPTDRELTIVTNSVPIASRVAAMPNVTLQ LLGGRVRGLTQAAVGEQALRVLDALRVDIAFIGTNAISLRHGLSTPDTDEAAVKRAMVSAANYVVVVADS SKMGQEDLIRFAPIDCVDTLITDAEIDPADRDGLTEQGVEVVIA*
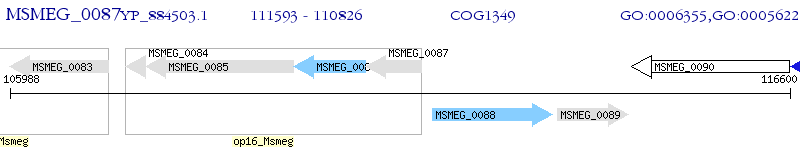
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0087 | - | - | 100% (255) | glucitol operon repressor |
| M. smegmatis MC2 155 | MSMEG_1377 | - | 7e-49 | 44.88% (254) | DeoR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_0509 | - | 1e-31 | 35.08% (248) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0750 | - | 1e-107 | 76.86% (255) | DeoR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3302 | - | 5e-34 | 35.80% (243) | PUTATIVE putative glucitol operon repressor |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0096 | - | 1e-110 | 80.39% (255) | DeoR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0750|M.gilvum_PYR-GCK MYPEERQQAIASLVMTKGRASVTELAHAYDVTTETVRRDLAVLDKAGIVR
Mvan_0096|M.vanbaalenii_PYR-1 MYPEERQQAIASLVMSKGRASVTELAQAYDVTTETVRRDLAVLDKAGVVR
MSMEG_0087|M.smegmatis_MC2_155 MYAEERQQAIAALVLSRGRASVAELAQAYDVTTETVRRDLAVLDRAGVVR
TH_3302|M.thermoresistible__bu LLASDRRREIFNRLVREGYVEAKELAEELGVDASTIRRDLDTLARSGQVK
: ..:*:: * :: .* ... ***. .* :.*:**** .* ::* *:
Mflv_0750|M.gilvum_PYR-GCK RVHGGAVPARSLHLVEPGVGERDVTRTEHKDAIAAAAVEFLPLSGATVLL
Mvan_0096|M.vanbaalenii_PYR-1 RVHGGAVPVRALHLVEPGVGERDVIRAEHKDAIAAAAAEFFPLSGASVLL
MSMEG_0087|M.smegmatis_MC2_155 RVHGGAVPVRTLHLVEAGVGERDSTRAEQKDAIAAAAAEFFPPTGSSVLL
TH_3302|M.thermoresistible__bu RTHGGARTVPGVSADLP-YSMKETARLDQKAAIAGVAVQRVR-DGDTVVL
*.**** .. : . . :: * ::* ***..*.: . * :*:*
Mflv_0750|M.gilvum_PYR-GCK DAGTTTMRIAGELPADRELVVVTNSVPIAARLAAMPSISLQLLGGRVRGV
Mvan_0096|M.vanbaalenii_PYR-1 DAGTTTMRIAAQLPADRELVIVTNSVPIAARLATVPSVSLQLLGGRVRGV
MSMEG_0087|M.smegmatis_MC2_155 DAGTTTARVAAHLPTDRELTIVTNSVPIASRVAAMPNVTLQLLGGRVRGL
TH_3302|M.thermoresistible__bu DSGSTTYEIAVRLRNHEDLTVITNDLRIATFIADLRRFRLLVAGGELLGS
*:*:** .:* .* ..:*.::**.: **: :* : . * : **.: *
Mflv_0750|M.gilvum_PYR-GCK TQAAVGEQTLRVLDTLRVDIAFIGTNAISARHGLSTPDSEEAAVKRAMVK
Mvan_0096|M.vanbaalenii_PYR-1 TQAAVGEQALRVLDTLRVDIAFIGTNAISVRHGLSTPDSEEAAVKRAMVR
MSMEG_0087|M.smegmatis_MC2_155 TQAAVGEQALRVLDALRVDIAFIGTNAISLRHGLSTPDTDEAAVKRAMVS
TH_3302|M.thermoresistible__bu VYTLTGRRTVEFLLDYAADWTFLGADAVDAAAGITNTNTLEIPVKRAMLE
. : .*.:::..* .* :*:*::*:. *::..:: * .*****:
Mflv_0750|M.gilvum_PYR-GCK AANYVVVAADSSKVGREDFVSFAPISSVDSLITDPEISPTDSAELTAHGV
Mvan_0096|M.vanbaalenii_PYR-1 AANYVVVAADSSKVGREDFVSFAPISSVDTLITDPEISAADSAEFGEHGV
MSMEG_0087|M.smegmatis_MC2_155 AANYVVVVADSSKMGQEDLIRFAPIDCVDTLITDAEIDPADRDGLTEQGV
TH_3302|M.thermoresistible__bu VAATTVVVADSSKFGHRALAKVADLDEVDEIITDSDLPSDSVDVFGKNLI
.* .**.*****.*:. : .* :. ** :***.:: . . : : :
Mflv_0750|M.gilvum_PYR-GCK EVICAGGRS-
Mvan_0096|M.vanbaalenii_PYR-1 EVICAGGRP-
MSMEG_0087|M.smegmatis_MC2_155 EVVIA-----
TH_3302|M.thermoresistible__bu TVADPAPTPG
* .