For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VNRKRLMLAAGVVALALPMAACTSSKPQADESSETPAAAGEAPAAAVTTVTAPPKASKNYNIAFLQGVVG DQFYITMQCGAQEEAANLGVTVNTQGPQKFDPTLQKPILDSIVASKPDALLVAPTDVQAMQMPLEQAAAA GIKVVLVDTTTNDPSYAVSAIASDNEGGGRAAFEAIKQLHPEGGKVMVMGLDPGISTTDARTKGFEEAVA EDPGFTYVGVQYSHNDPATAAQLIGAQLQRDPDLVGVFAANLFTAEGSATGIKQAGKSEQVAVVGFDAGP NQIQALREGTVQALVAQDPGTIGKFGVDEAVTALEGGENSPNVQTGFTIITRENLDGEGGAAAYKSSC
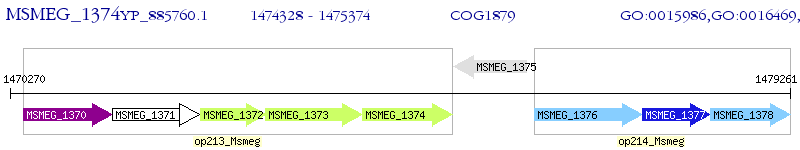
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1374 | - | - | 100% (348) | ribose ABC transporter, periplasmic binding protein |
| M. smegmatis MC2 155 | MSMEG_3095 | - | 4e-19 | 27.15% (291) | D-ribose-binding periplasmic protein |
| M. smegmatis MC2 155 | MSMEG_6804 | - | 3e-15 | 23.47% (311) | sugar ABC transporter substrate-binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_4578 | - | 9e-11 | 22.91% (275) | ABC sugar transporter, periplasmic ligand binding protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00398 | - | 5e-09 | 25.23% (218) | putative D-ribose-binding protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4011 | - | 6e-11 | 23.75% (341) | periplasmic binding protein/LacI transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1374|M.smegmatis_MC2_155 -MNRKRLMLAAGVVALALPMAACTSSKPQADESSETPAAAGEAPAAAVTT
Mvan_4011|M.vanbaalenii_PYR-1 -MTFKRLAAFAGAGVLAMGIVSCSSTGGAPEQSGDGGGGG---------T
MLBr_00398|M.leprae_Br4923 MSTGTSVPLRVASIMLILGLLPAILPACGFSESHHKLTIG----------
Mflv_4578|M.gilvum_PYR-GCK -MRRSRTALLGAVGVTMSLVAASAGAANASPDAGDVTMVN----------
. . : .. . :: .
MSMEG_1374|M.smegmatis_MC2_155 VTAPPKASKNYNIAFLQGVVGDQFYITMQCGAQEEAANLGVTVNTQGPQK
Mvan_4011|M.vanbaalenii_PYR-1 VDTP----RMTIAMITHEVPGDSFWDLVRKGAEAAAKKDNIELRYSNDP-
MLBr_00398|M.leprae_Br4923 --------------VSYPTANSPFWNAYTRFIDEGSHQLGVYINAVSAEE
Mflv_4578|M.gilvum_PYR-GCK ---------------VVKVEGIPWFNRMAVGVENFASRTGVHASQVGADD
. . :: : : . .: .
MSMEG_1374|M.smegmatis_MC2_155 FDPTLQKPILDSIVASKPDALLVAPTDVQAMQMPLEQAAAAGIKVVLVDT
Mvan_4011|M.vanbaalenii_PYR-1 -EAPNQANLVQSAIDSNVDGIAVTLAKPDAMQAAVKSAESKGIPVVAFNA
MLBr_00398|M.leprae_Br4923 -DEQKQLSDVETLINQGIDGLIVTPQSTSIAPTLLRIATQANIPVVVVDR
Mflv_4578|M.gilvum_PYR-GCK TSAQKQVQLIADLIPRRPTAITVVPNSVDALEDVLGQARAQGITVVSHEA
. * : : .: *. . . : * .* ** :
MSMEG_1374|M.smegmatis_MC2_155 TT---NDPSYAVSAIASDNEGGGRAAFEAIKQLHPEGG-KVMVMGLDPGI
Mvan_4011|M.vanbaalenii_PYR-1 G----MDSWKAMGVKEYFGQDGRIAGQGVGDRLRAEGATKSICVIQEQGH
MLBr_00398|M.leprae_Br4923 YPGYAPGQNKNADYVAFLGPNNEKAGSGIAEALMAGGGSKFLALGGMPGN
Mflv_4578|M.gilvum_PYR-GCK TG----IRNADIDIEAFDNAAFGRQTMQSLAQCMEGQGEYVQFVGSVAMT
. . . :
MSMEG_1374|M.smegmatis_MC2_155 STTDARTKGFEEAVAED----PGFTYVGVQYSHNDPATAAQLIGAQLQRD
Mvan_4011|M.vanbaalenii_PYR-1 VDLEARCAGVKETFPAT----EILNVNG-----KDMPSVESTITAKLQQD
MLBr_00398|M.leprae_Br4923 SVAQGRKAGLESALTAVGHRLVQFQYAG---DSEDKGLAAAENMLQAHPA
Mflv_4578|M.gilvum_PYR-GCK THMQWVDAAHDMQLAEFP---SMTRIEGPVETANDEPTAYARATELLVKY
: . . .. * :* .
MSMEG_1374|M.smegmatis_MC2_155 PDLVGVFAANLFTAEGSATGIKQAGKSEQVAVVGFDAGPNQIQALREGTV
Mvan_4011|M.vanbaalenii_PYR-1 PSIDYVVTLGAPFALTAVQSAKNAGSSAKIG--TFDTNAALVEAIQNGDV
MLBr_00398|M.leprae_Br4923 GDANAIWCFNDKLCQGAIKAVYNANREKEFIFGGMDLTPQAIAAIENGLY
Mflv_4578|M.gilvum_PYR-GCK PNLKGFQGAASTDVPGIARAVRDAGLQDRTCVMGTAVPSVASTFFADGSI
. . . :*. . . . : :*
MSMEG_1374|M.smegmatis_MC2_155 QALVAQDPGTIGKFGVDEAVTALEGG---ENSPNVQTGFTIITRENLDGE
Mvan_4011|M.vanbaalenii_PYR-1 QWAVDQQPYLQGYLAVDSLWLYLSNGNVIGGGQPTLTGPAFIDKSNIDAV
MLBr_00398|M.leprae_Br4923 TVSLG-AHWLEGGFGLAILYRKIHGQD--PAERMVKLDLLKVDKSNVAKF
Mflv_4578|M.gilvum_PYR-GCK DKIFFWDPAMAAEAQLQIALMLVQGEKIETGTDLNVPGYDSLTK--LDGY
. . : : . . : : :
MSMEG_1374|M.smegmatis_MC2_155 GGAAAYKSSC----------------
Mvan_4011|M.vanbaalenii_PYR-1 AEYAKGGTR-----------------
MLBr_00398|M.leprae_Br4923 KARYIDNTPHYNWKSTTPFDMTITLN
Mflv_4578|M.gilvum_PYR-GCK DNVFVGNAALEADANTISQNQF----
: