For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PTPFPAEFRADVIAVARKGEAPLRQIAKDFGISEACLHRWLKIADREDGRGQPASPADADDMAAQLREAH KRIKLLEQEAEVMRRAVGYLSRDANPK*
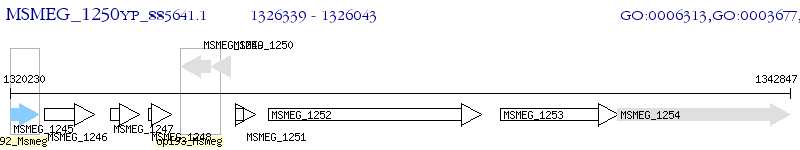
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1250 | - | - | 100% (98) | ISMsm7, transposase orfA |
| M. smegmatis MC2 155 | MSMEG_6889 | - | 9e-19 | 48.91% (92) | transposase |
| M. smegmatis MC2 155 | MSMEG_4106 | - | 2e-17 | 47.83% (92) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_3194 | - | 1e-17 | 47.83% (92) | transposase IS3/IS911 family protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2087 | - | 5e-17 | 45.26% (95) | transposase |
| M. marinum M | MMAR_3793 | - | 2e-16 | 42.42% (99) | transposase |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5960 | - | 5e-17 | 45.65% (92) | transposase IS3/IS911 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2087|M.abscessus_ATCC_1997 MTSELACPKRAGWKDVSVPRPYPREFRDDVVRVARNRDDGVTIEQIATDF
Mvan_5960|M.vanbaalenii_PYR-1 -----------------MARPYPREFRDDVVRVARNRDDGVTIEQIATDF
Mflv_3194|M.gilvum_PYR-GCK -----------------MARPYPREFRDDVVRVARNRDDGVTIEQIATDF
MMAR_3793|M.marinum_M -----------------MSKPYPGEFRRDVVRVARERGPGVSVEQVARDF
MSMEG_1250|M.smegmatis_MC2_155 -----------------MPTPFPAEFRADVIAVARKGE--APLRQIAKDF
:. *:* *** **: ***: ..:.*:* **
MAB_2087|M.abscessus_ATCC_1997 GVHPMTLTKWMRQADVDEG----AKPGKSTNDSADLRELRRRNRLLEQEN
Mvan_5960|M.vanbaalenii_PYR-1 GVHPMTLHKWMRQADIDEG----AKPGKSTGESAELRELRRRNRLLEQEN
Mflv_3194|M.gilvum_PYR-GCK GVHPMTLQKWLRQADIEEG----TKPGKSASESGELREARRRIKLLEQEN
MMAR_3793|M.marinum_M GIHQATLNAWLRAADADEG----PKSIAEPDVLRENRDLKRRNRLLEQEN
MSMEG_1250|M.smegmatis_MC2_155 GISEACLHRWLKIADREDGRGQPASPADADDMAAQLREAHKRIKLLEQEA
*: * *:: ** ::* ... . : *: ::* :*****
MAB_2087|M.abscessus_ATCC_1997 EVLRRAAAYLSQANLPGKG------STRS--
Mvan_5960|M.vanbaalenii_PYR-1 EVLRRAAAYLSQANLPGKG------STRS--
Mflv_3194|M.gilvum_PYR-GCK EVLRRAAAYLSQANLPGKG------STRS--
MMAR_3793|M.marinum_M EVLRKAAAYLGRGSLPGKGRFYGLVSSGGGL
MSMEG_1250|M.smegmatis_MC2_155 EVMRRAVGYLSRDANPK--------------
**:*:*..**.: *