For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
LAPCPAAVDDEFVVNKNSSRTLAPLANDTDAKGNSMIDPETVTIVGQPSHGTVTVNDNGAVTDTSTIGAA S
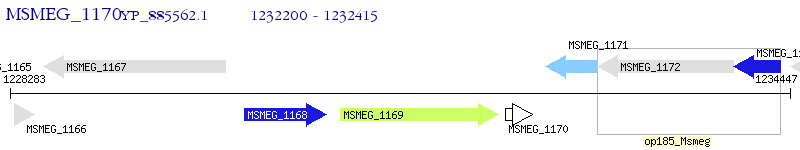
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1170 | - | - | 100% (71) | hypothetical protein MSMEG_1170 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0506 | - | 5e-11 | 48.39% (62) | putative outer membrane adhesin like protein |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK MGVSRCIGRVGALAVALGVGTAVTFPHTALADEGAGSNGSVSSSSRESTS
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GATGTSDRESSSSPSKPAATRPSDSDDTETSGPDGDEPNADGSEPTEEDT
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK EPSSTPATSPEPPADPVVEDTTDEALADPDPAESLAPSTLHPESGSTPEP
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VHSITATTVADLPTSAPPEEPASQPPRTDVPAAYPRTSPAIQAPASSVGA
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK LSTPVSQPVSLVGALVSTVLAPFSGGAPAAPSQPPLLWVVLAWVRRELQR
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK TFANSTPTTTPQSVTLTLDSATSVSAPIEFDADDVDDDPLVYTVPGRGAV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GGPTHGTVTVDQTTGTFTYNPDDAYALIGGTDTFTYTVSDAGARRTFFGI
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK PLSRSATTATGIVTVTLNPVHVAPVTNGDTATTVEDRPVTIAVLGNDTDA
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK NGDALTVSGVTQGTSGTVTFTASGVTYTPNADFTGTDTFTYTATDGRSAG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK TATVTVTVTPVDDAPVAVGDSVTVAEDSGTTVIDVLANDIDVDGGPKVVT
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GVTPPAHGTVTAIGPSLSYTPAADFDGTDAFTYTLNDGSTATVTVIVTPV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK DDKPVAVGDTVTVAEDSSPTVIDVLANDTDIDAGPKTITGVSQPAKGNVV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VTGTTVIYTPTADFTGTDTFTYTLNGGATATVTVTVTPVDDAPVTVGDTA
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK TVAEDSGPTIIDVLANDTDIDAGPIAITAVTQPVDGTVTFTASEVTYTPG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK ADFHGTDTFTYTLNGGATATVTVTVTPVDDAPVTVGDAVTVAEDSGPTII
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK DVLVNDTDIDAGPIAITAVTQPVDGTVTFTASEVTYTPGADFHGTDTFTY
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK TLNGGSVAVVTVIVDRVDDTPIAFGDSVTVAEDSGPTTIDVLANDTDIDN
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GPIAITAVTQPTNGTIDFTVSRVTYTPGANFHGTDTFTYTLNGGATATVT
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VTVNAAPVIGQVTTTPGVGNTWEVSVSADDSENDTLTTTVTSADPTVPLT
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VTKQTDGTFQVVADPTWASAHPAAQIAVTVTVTDTYGSSATITTAIGTAN
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK NVISLGWDATGVGNVPALPAGVSYTQVAGGAFHTVFLRSDGSVVGAGQNN
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK YGQLDIPPLPTGITYTQVAAGAWHTVLLRSDGTVVAVGDNFKGQIDIPEP
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GLGVSYTGVAAGELGTILLRSDGTAVAVGDGQYGQLEIPTLPPGVRYTQA
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK AIGSVRSALLRSDGTVVTVGMNDAVPTLPAGVRYISVASGAGHAVLLRSD
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GVAVAFGANDWGQTDIPALPAGVVYVNIAASRYHSVLLRSDGTSVAAGQN
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK TSEQLDIPVLPVGVTYTGGAAGGYSTWLISSTTATPFNTPPVAVDDAMTV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK AEGNTATISVTANDTDADGTIDTATVVITRQSTAGSVAVNAAGTVTYVSN
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GTEVTADSFAYRIDDALGATSNEATVFITITPVDDAPVAVNDAVTVAEGG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK TITIAVIGNDTDADGTIDTTTVVIVTQPTAGTATVNGDGTVTYVSDGSEV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK TSDSFTYTVKDTAGLTSNVATVSITVNPVNDAPVAVNDAYTVAENGLLVI
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK DAPGLLGNDLDSEHPFIYITTITNPRHGTFDGILSDGSFTYRPHAGYFGT
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK DSFTYQINDVTGLTSNTATVTITITPVDDAPVAVNDAVTVAEDSGTTVID
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VLANDTDIDAGPKTITSITQPNSGTTTITATGVAYTPNENFHGTDTFTYT
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK LNGGTSAEVTVTVTPVNDAPVAPDKVVTISEDTWSVAIHLTDGATDVDGD
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK PLRRGYVTYPAIGSTFVDSDNLGIQRLLRYYPPANYTGQITIPYTVWDGQ
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VFSNWATITINVTPVNDVPVAVNDSAAVDAGGTTTITLTANDTDVDTDNG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VDPATIVIGTQPSAGTVTVNSDGTVTYTSTGTTTTTDIFTYTVEDTTGLV
MSMEG_1170|M.smegmatis_MC2_155 ------------MAPCPAAVDDEFVVNKNSSRTLAPLANDTDAKGNSMID
Mflv_0506|M.gilvum_PYR-GCK SNPATVTVNVTATATPPVAVNDTVTVDEGGTTTLAVIANDTDADGD--ID
*. *.**:* ..*::..: *** :******.*: **
MSMEG_1170|M.smegmatis_MC2_155 PETVTIVGQPSHGTVTVNDNG-----------------------------
Mflv_0506|M.gilvum_PYR-GCK TATVVIVRQPTAGTATVNSDGTVSYASDGSEITTDSFEYTVRDVAGAVSN
. **.** **: **.***.:*
MSMEG_1170|M.smegmatis_MC2_155 ----------------AVTDTSTIGAAS----------------------
Mflv_0506|M.gilvum_PYR-GCK AATVIITVTPVDDAPVAVNDTVTITEDTLATFIDVLANDTDIDGGPKTIT
**.** ** :
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK SITSPAGGHALVIGNRVRYFSAPNFHGTETFTYTLNGGATATVTVIVTPI
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK SEAPVIHRVTSTPGVGNTWVLSVDASDDDGGALTTTVTSANATVPLTITR
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK LPGGAIQVDIDPTWARAHPGAQLPVTVTVTDAENISATKQLTIGTVSNAV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK FFGHNIYGQRNIPALPEGVTYTQVATASLSHTVLLRSDGTAVAVGDNFRG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK QLNIPPLPTGMTYTQVANGTEHTVLLRSDGTAIAVGNNSDGQLNIPRLPA
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GVTYTRIAAGNYGTVLLRSDGTAVGAGYNTYGQLNIPQPPPGVTYTQAAV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GLTHTVLLRSDGTAVALGDNAFGQTVIPALPTGLKYTQVAAGDLHSVLLR
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK SDGSVVAFGNNQWGQSTVPALPAGLRYTQVAAGRYNSMLLRSDGTAIGFG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK QNASSQLNIPVLPAGLVYTQAAASNHTVLLFSTSSSFNSLPLATNDAATV
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK AEGGTITFSVTANDTDTDGTLNTATVVITRQPTSGSVTVNAGGTVTYVSN
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK GAEVSADSFAYRVNDNKGATSNEATVSITITPVNDAPVAVDDAYTVAKNG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VLVISAPGLLANDSDPEGNSFTIVAATGTQHGTVDPLYSNGSFTYRPAAG
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK YSGTDTFTYTISDGTSTAQATVTIAVSAVDNQAPVAVADSYTVAEDGVLN
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK VTGPGVLANDTDADGDPLSVVAAGPTSHGTFTLRSDGSFTYTPVANFHGT
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK DSFTYQVSDGSGPSAPATVTITVTPVNDAPVVQNDTIPFPKDGFIGFSLA
MSMEG_1170|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0506|M.gilvum_PYR-GCK NRSSDVDGDTLTATLISGTTHGTLFFDGTNRAFSYRPNTGFTGQDSFTYK
MSMEG_1170|M.smegmatis_MC2_155 ------------------
Mflv_0506|M.gilvum_PYR-GCK VNDGTVDSAIATVTLNVS