For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDEAGEARGAQELGGEASAEVAVTDLQAEQTPPPSAPPPDLTAAAFFDVDNTLVQGSSLVHFARGLAARK YFTYRDILGIAYAQAKFQLTGKENSDDVAAGRRKALAFIEGRSTAELVALGEEIYDEIIADKIWPGTRAL AQMHLDAGQQVWLVTATPYELAATIAKRLGFTGALGTVAESVDGVFTGRLVGDILHGTGKAHAVRSLAIR EGLNLRRCTAYSDSFNDVPMLSLVGTAVAINPDAALRDVARERGWEIRDFRTARKAARIGVPSALALGAL GGALAAVASRRHDLR*
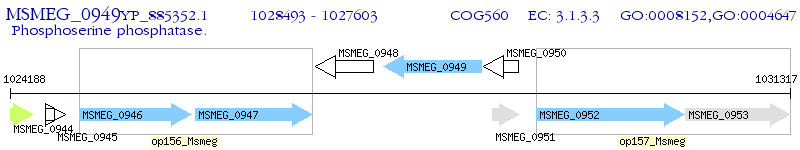
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0949 | - | - | 100% (296) | HAD-superfamily protein subfamily protein IB hydrolase |
| M. smegmatis MC2 155 | MSMEG_6173 | - | 7e-29 | 33.33% (261) | morphological differentiation-associated protein |
| M. smegmatis MC2 155 | MSMEG_4704 | - | 9e-25 | 30.97% (226) | acyltransferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0517c | serB1 | 1e-129 | 81.36% (295) | phosphoserine phosphatase |
| M. gilvum PYR-GCK | Mflv_0069 | - | 1e-138 | 87.02% (285) | HAD family hydrolase |
| M. tuberculosis H37Rv | Rv0505c | serB1 | 1e-129 | 81.36% (295) | phosphoserine phosphatase |
| M. leprae Br4923 | MLBr_02424 | - | 1e-123 | 79.86% (283) | hypothetical protein MLBr_02424 |
| M. abscessus ATCC 19977 | MAB_3995 | - | 1e-122 | 81.39% (274) | hypothetical protein MAB_3995 |
| M. marinum M | MMAR_0833 | serB1 | 1e-129 | 83.39% (283) | phosphoserine phosphatase SerB1 |
| M. avium 104 | MAV_4645 | - | 1e-130 | 83.80% (284) | HAD-superfamily protein subfamily protein IB hydrolase |
| M. thermoresistible (build 8) | TH_3424 | - | 1e-129 | 80.55% (293) | PUTATIVE HAD-superfamily subfamily IB hydrolase, TIGR01490 |
| M. ulcerans Agy99 | MUL_4577 | serB1 | 1e-129 | 83.04% (283) | phosphoserine phosphatase SerB1 |
| M. vanbaalenii PYR-1 | Mvan_0843 | - | 1e-138 | 86.69% (293) | HAD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0517c|M.bovis_AF2122/97 MGLTCWPRTAAGRVHDESRCGLANFDTALGLQINPRQPRAPPRICRIGLI
Rv0505c|M.tuberculosis_H37Rv MGLTCWPRTAAGRVHDESRCGLANFDTALGLQINPRQPRAPPRICRIGLI
MMAR_0833|M.marinum_M --------------------------------------------------
MUL_4577|M.ulcerans_Agy99 --------------------------------------------------
MAV_4645|M.avium_104 --------------------------------------------------
MLBr_02424|M.leprae_Br4923 --------------------------------------------------
MSMEG_0949|M.smegmatis_MC2_155 --------------------------------------------------
TH_3424|M.thermoresistible__bu --------------------------------------------------
Mflv_0069|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0843|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_3995|M.abscessus_ATCC_1997 --------------------------------------------------
Mb0517c|M.bovis_AF2122/97 TAAASATGQAPRLGVMMVSSHLGSPDQAGHVDLASPADPPPPDASASHSP
Rv0505c|M.tuberculosis_H37Rv TAAASATGQAPRLGVMMVSSHLGSPDQAGHVDLASPADPPPPDASASHSP
MMAR_0833|M.marinum_M ----------------MASSDLGSSSGGGHFDLESLAEN----ASATRAL
MUL_4577|M.ulcerans_Agy99 ----------------MASSDLGSSSGGGHFDLESLAEN----ASATRAL
MAV_4645|M.avium_104 ----------------MTSSDPVNADPTGHVDLESLAAD----ASAARAI
MLBr_02424|M.leprae_Br4923 ----------------MASPDLSNAYN-GRIDLGSLANN----ASINRAL
MSMEG_0949|M.smegmatis_MC2_155 -----------------MTDEAGEARGAQELGGEASAE--------VAVT
TH_3424|M.thermoresistible__bu --------VAESRGVETTPTDASHGPREQETAGKASAE--------AAVT
Mflv_0069|M.gilvum_PYR-GCK -------------MSDIGG-------RAQQLAGEASAE--------AAVE
Mvan_0843|M.vanbaalenii_PYR-1 -------------MSDTGGIEIGSGSRAQELAGEVSAE--------VAAE
MAB_3995|M.abscessus_ATCC_1997 -----------MTSPQINGEPPAEGPTEEQVLAEAFAED------VARAA
. *
Mb0517c|M.bovis_AF2122/97 VDMPAPVAAAGSDRQPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARHY
Rv0505c|M.tuberculosis_H37Rv VDMPAPVAAAGSDRQPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARHY
MMAR_0833|M.marinum_M ADMTVPTEGSAADPQPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARHY
MUL_4577|M.ulcerans_Agy99 ADMTVPTEGSAADPQPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARHY
MAV_4645|M.avium_104 EGMQEAAATQEDRPQPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLATRNY
MLBr_02424|M.leprae_Br4923 NDMPTAVDDAGVRPQPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARDY
MSMEG_0949|M.smegmatis_MC2_155 DLQAEQ---TPPPSAPPPDLTAAAFFDVDNTLVQGSSLVHFARGLAARKY
TH_3424|M.thermoresistible__bu ELAPE-----PPPPAPPPDLTAAAFFDVDNTLVHGSSLVHFARGLAARNY
Mflv_0069|M.gilvum_PYR-GCK GLTEPLGPAAPPPPPPPPDLTAAAFFDVDNTLVHGSSLVHFARGLAAREY
Mvan_0843|M.vanbaalenii_PYR-1 GLAQPLDAVAAPPPPPP-DLTAAAFFDVDNTLVHGSSLVHFARGLAAREY
MAB_3995|M.abscessus_ATCC_1997 SFAEADAAAEQPPGPPPADLTAAAFFDVDNTLVHGSSLVHFGRGLAQRDY
** ***************:*** ***.**** *.*
Mb0517c|M.bovis_AF2122/97 FTYRDVLGFLYAQAKFQLLGKENSNDVAAGRRKALAFIEGRSVAELVALG
Rv0505c|M.tuberculosis_H37Rv FTYRDVLGFLYAQAKFQLLGKENSNDVAAGRRKALAFIEGRSVAELVALG
MMAR_0833|M.marinum_M FTYRDVLGFVYAQAKFQLLGTENSEDVAAGRRKALAFIEGRSVDELLNLG
MUL_4577|M.ulcerans_Agy99 FTYRDVLGFVYAQAKFQLLGTENSEDVAAGRRKALAFIEGRSVDELLNLG
MAV_4645|M.avium_104 FTYRDVLGFIYAQAKFQVLGKENSNDVAAGRRKALAFIEGRSVEQLVALG
MLBr_02424|M.leprae_Br4923 FTYRDVLGFIYAQAKFQLLGKENSQDVAAGQRKALAFIEGRSVEQLVALG
MSMEG_0949|M.smegmatis_MC2_155 FTYRDILGIAYAQAKFQLTGKENSDDVAAGRRKALAFIEGRSTAELVALG
TH_3424|M.thermoresistible__bu FSYRDMLRFVYAQAKFQLTGRENSDDVAAGRRKALSFIEGRSVAELIALG
Mflv_0069|M.gilvum_PYR-GCK FTYQDLARFAYAQAKFQLTGRENSDDVAAGRRKALSFIEGRSTAELVALG
Mvan_0843|M.vanbaalenii_PYR-1 FTYQDLARFALAQAKFQLTGRENSGDVAAGRRKALAFIEGRSTAELVALG
MAB_3995|M.abscessus_ATCC_1997 FQYSDMLQFVWAQAKFRLTGRENSDDVAAGRQKALSFIEGRSVDELVALS
* * *: : *****:: * *** *****::***:******. :*: *.
Mb0517c|M.bovis_AF2122/97 EEIYDEIIADKIWDGTRELTQMHLDAGQQVWLITATPYELAATIARRLGL
Rv0505c|M.tuberculosis_H37Rv EEIYDEIIADKIWDGTRELTQMHLDAGQQVWLITATPYELAATIARRLGL
MMAR_0833|M.marinum_M EEIYDEIIADKIWPGTRELAQMHLDAGQQVWLITATPYELAATIARRLGL
MUL_4577|M.ulcerans_Agy99 EEIYDDIIADKIWPGTRELAQMHLDAGQQVWLITATPYELAATIARRLGL
MAV_4645|M.avium_104 EEIYDEIIADKIWPGTRELTQMHLDAGQQVWLITATPYELAATIARRLGL
MLBr_02424|M.leprae_Br4923 EEIYDEIIADKIWAGTRQLTQIHLDAGQQVWLITATPYELAATIARRLGL
MSMEG_0949|M.smegmatis_MC2_155 EEIYDEIIADKIWPGTRALAQMHLDAGQQVWLVTATPYELAATIAKRLGF
TH_3424|M.thermoresistible__bu EEIYDEIIADKIWPGTLALAQMHLDAGQQVWLVTATPYELAATIARRLGF
Mflv_0069|M.gilvum_PYR-GCK EDIYDEIIADKIWPGTRRLAQMHLDAGQQVWLVTATPYELAETIARRLGL
Mvan_0843|M.vanbaalenii_PYR-1 EEIYDEIIADKIWPGTRALAQMHLDAGQQVWLVTATPYELADTIARRLGL
MAB_3995|M.abscessus_ATCC_1997 EEIYDETIADKIWPGTRALTQMHLDAGQQVWLVTATPRELAETIARRLGL
*:***: ****** ** *:*:**********:**** *** ***:***:
Mb0517c|M.bovis_AF2122/97 TGALGTVAESVDGIFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTAY
Rv0505c|M.tuberculosis_H37Rv TGALGTVAESVDGIFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTAY
MMAR_0833|M.marinum_M TGALGTVAESVDGVFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTAY
MUL_4577|M.ulcerans_Agy99 TGALGTVAESVDGVFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTAY
MAV_4645|M.avium_104 TGALGTVAESVDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLKRCTAY
MLBr_02424|M.leprae_Br4923 TGALGTVAESVDGIFTGRLVDELLHGVGKAHAVRSLAIREGLNLKRCTAY
MSMEG_0949|M.smegmatis_MC2_155 TGALGTVAESVDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLRRCTAY
TH_3424|M.thermoresistible__bu TGALGTVAESVDGVFTGRLVGDILHGVGKAHAVRNLAVREGLNLRRCTAY
Mflv_0069|M.gilvum_PYR-GCK TGALGTVAESIDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLRRCTAY
Mvan_0843|M.vanbaalenii_PYR-1 TGALGTVAESIDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLRRCTAY
MAB_3995|M.abscessus_ATCC_1997 TGALGTVAESVDGVFTGRLVGDILHGPGKARAVRNLAIRNGLNLKRCTAY
**********:**:******.::*** ***:***.**:*:****:*****
Mb0517c|M.bovis_AF2122/97 SDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRIARKAARIGV
Rv0505c|M.tuberculosis_H37Rv SDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRIARKAARIGV
MMAR_0833|M.marinum_M SDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRTARKAARIGV
MUL_4577|M.ulcerans_Agy99 SDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRTARKAARIGV
MAV_4645|M.avium_104 SDSYNDVPMLSLVGTAVAINPDARLRALARERGWEIRDFRTARKAARIGV
MLBr_02424|M.leprae_Br4923 SDSYNDVPMLSLVGTAVAINPDAQLRSLARERGWEIRDFRTARKAARIGV
MSMEG_0949|M.smegmatis_MC2_155 SDSFNDVPMLSLVGTAVAINPDAALRDVARERGWEIRDFRTARKAARIGV
TH_3424|M.thermoresistible__bu SDSYNDVPMLSLVGTAVAINPDAALRSLARERGWEIRDFRTARKAARXXX
Mflv_0069|M.gilvum_PYR-GCK SDSFNDVPMLSLVGTAVAINPDAALRDLARQRGWEVRDFRTARKAARIGV
Mvan_0843|M.vanbaalenii_PYR-1 SDSFNDVPMLSLVGTAVAINPDADLRDLARERGWEIRDFRTARKAARIGV
MAB_3995|M.abscessus_ATCC_1997 SDSVNDVPMLSLVGTAVAINPDAELRDVARRRGWEVRDFRTARKAARIGV
*** ******************* ** :**.****:**** ******
Mb0517c|M.bovis_AF2122/97 PSALALGAAGGALAALASRRQSR--------------
Rv0505c|M.tuberculosis_H37Rv PSALALGAAGGALAALASRRQSR--------------
MMAR_0833|M.marinum_M PSALALGAAGGALAALASRRQSR--------------
MUL_4577|M.ulcerans_Agy99 PSALALGAAGGALAALASRRQSR--------------
MAV_4645|M.avium_104 PSALALGAAGGALAALASRRQSR--------------
MLBr_02424|M.leprae_Br4923 PSALALG---GALAAAVSRRRDRE-------------
MSMEG_0949|M.smegmatis_MC2_155 PSALALGALGGALAAVASRRHDLR-------------
TH_3424|M.thermoresistible__bu SAARCSAAGRSASTRTWCRRWPTRSPRWCAAASRWRW
Mflv_0069|M.gilvum_PYR-GCK PSALALGAVGGALAAAVSHRDKK--------------
Mvan_0843|M.vanbaalenii_PYR-1 PSALALGAVGGALAAAVSRREKK--------------
MAB_3995|M.abscessus_ATCC_1997 PSAVLLGAAGGALAAALARREK---------------
.:* . .* : .:*