For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ASSDLGSSSGGGHFDLESLAENASATRALADMTVPTEGSAADPQPPIDLTAAAFFDVDNTLVQGSSAVHF GRGLAARHYFTYRDVLGFVYAQAKFQLLGTENSEDVAAGRRKALAFIEGRSVDELLNLGEEIYDEIIADK IWPGTRELAQMHLDAGQQVWLITATPYELAATIARRLGLTGALGTVAESVDGVFTGRLVGEILHGTGKAH AVRSLAIREGLNLKRCTAYSDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRTARKAARIGVP SALALGAAGGALAALASRRQSR*
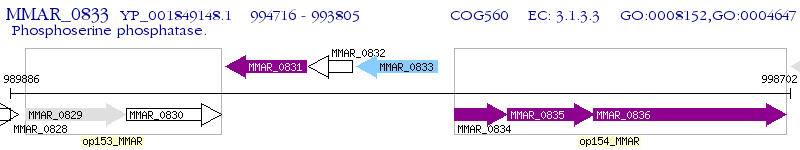
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_0833 | serB1 | - | 100% (303) | phosphoserine phosphatase SerB1 |
| M. marinum M | MMAR_5149 | serB | 4e-33 | 33.82% (272) | phosphoserine phosphatase SerB |
| M. marinum M | MMAR_3834 | plsC | 1e-24 | 29.93% (294) | bifunctional transmembrane phospholipid biosynthesis enzyme |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0517c | serB1 | 1e-147 | 87.30% (307) | phosphoserine phosphatase |
| M. gilvum PYR-GCK | Mflv_0069 | - | 1e-128 | 80.49% (287) | HAD family hydrolase |
| M. tuberculosis H37Rv | Rv0505c | serB1 | 1e-147 | 87.30% (307) | phosphoserine phosphatase |
| M. leprae Br4923 | MLBr_02424 | - | 1e-140 | 84.16% (303) | hypothetical protein MLBr_02424 |
| M. abscessus ATCC 19977 | MAB_3995 | - | 1e-120 | 75.08% (297) | hypothetical protein MAB_3995 |
| M. avium 104 | MAV_4645 | - | 1e-146 | 87.46% (303) | HAD-superfamily protein subfamily protein IB hydrolase |
| M. smegmatis MC2 155 | MSMEG_0949 | - | 1e-129 | 83.39% (283) | HAD-superfamily protein subfamily protein IB hydrolase |
| M. thermoresistible (build 8) | TH_3424 | - | 1e-118 | 77.03% (283) | PUTATIVE HAD-superfamily subfamily IB hydrolase, TIGR01490 |
| M. ulcerans Agy99 | MUL_4577 | serB1 | 1e-169 | 99.67% (303) | phosphoserine phosphatase SerB1 |
| M. vanbaalenii PYR-1 | Mvan_0843 | - | 1e-129 | 79.33% (300) | HAD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0833|M.marinum_M --------------------------------------------------
MUL_4577|M.ulcerans_Agy99 --------------------------------------------------
Mb0517c|M.bovis_AF2122/97 MGLTCWPRTAAGRVHDESRCGLANFDTALGLQINPRQPRAPPRICRIGLI
Rv0505c|M.tuberculosis_H37Rv MGLTCWPRTAAGRVHDESRCGLANFDTALGLQINPRQPRAPPRICRIGLI
MAV_4645|M.avium_104 --------------------------------------------------
MLBr_02424|M.leprae_Br4923 --------------------------------------------------
Mflv_0069|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0843|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_0949|M.smegmatis_MC2_155 --------------------------------------------------
TH_3424|M.thermoresistible__bu --------------------------------------------------
MAB_3995|M.abscessus_ATCC_1997 --------------------------------------------------
MMAR_0833|M.marinum_M ----------------MASSDLGSSSGGGHFDLESLAEN----ASATRAL
MUL_4577|M.ulcerans_Agy99 ----------------MASSDLGSSSGGGHFDLESLAEN----ASATRAL
Mb0517c|M.bovis_AF2122/97 TAAASATGQAPRLGVMMVSSHLGSPDQAGHVDLASPADPPPPDASASHSP
Rv0505c|M.tuberculosis_H37Rv TAAASATGQAPRLGVMMVSSHLGSPDQAGHVDLASPADPPPPDASASHSP
MAV_4645|M.avium_104 ----------------MTSSDPVNADPTGHVDLESLAAD----ASAARAI
MLBr_02424|M.leprae_Br4923 ----------------MASPDLSNAYN-GRIDLGSLANN----ASINRAL
Mflv_0069|M.gilvum_PYR-GCK -----------------MSDIGG-------RAQQLAGEA---SAE--AAV
Mvan_0843|M.vanbaalenii_PYR-1 -----------------MSDTGGIEIGSGSRAQELAGEV---SAE--VAA
MSMEG_0949|M.smegmatis_MC2_155 -----------------MTDEAG----EARGAQELGGEA---SAE--VAV
TH_3424|M.thermoresistible__bu --------VAESRGVETTPTDAS----HGPREQETAGKA---SAE--AAV
MAB_3995|M.abscessus_ATCC_1997 ---------------MTSPQINGEPPAEGPTEEQVLAEA---FAEDVARA
. . *.
MMAR_0833|M.marinum_M ADMTVPTEGSAADP-QPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARH
MUL_4577|M.ulcerans_Agy99 ADMTVPTEGSAADP-QPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARH
Mb0517c|M.bovis_AF2122/97 VDMPAPVAAAGSDR-QPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARH
Rv0505c|M.tuberculosis_H37Rv VDMPAPVAAAGSDR-QPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARH
MAV_4645|M.avium_104 EGMQEAAATQEDRP-QPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLATRN
MLBr_02424|M.leprae_Br4923 NDMPTAVDDAGVRP-QPPIDLTAAAFFDVDNTLVQGSSAVHFGRGLAARD
Mflv_0069|M.gilvum_PYR-GCK EGLTEPLGPAAPPPPPPPPDLTAAAFFDVDNTLVHGSSLVHFARGLAARE
Mvan_0843|M.vanbaalenii_PYR-1 EGLAQPLDAVAAPPPPPP-DLTAAAFFDVDNTLVHGSSLVHFARGLAARE
MSMEG_0949|M.smegmatis_MC2_155 TDLQAEQ---TPPPSAPPPDLTAAAFFDVDNTLVQGSSLVHFARGLAARK
TH_3424|M.thermoresistible__bu TELAPE-----PPPPAPPPDLTAAAFFDVDNTLVHGSSLVHFARGLAARN
MAB_3995|M.abscessus_ATCC_1997 ASFAEADAAAEQPPGPPPADLTAAAFFDVDNTLVHGSSLVHFGRGLAQRD
: ** ***************:*** ***.**** *.
MMAR_0833|M.marinum_M YFTYRDVLGFVYAQAKFQLLGTENSEDVAAGRRKALAFIEGRSVDELLNL
MUL_4577|M.ulcerans_Agy99 YFTYRDVLGFVYAQAKFQLLGTENSEDVAAGRRKALAFIEGRSVDELLNL
Mb0517c|M.bovis_AF2122/97 YFTYRDVLGFLYAQAKFQLLGKENSNDVAAGRRKALAFIEGRSVAELVAL
Rv0505c|M.tuberculosis_H37Rv YFTYRDVLGFLYAQAKFQLLGKENSNDVAAGRRKALAFIEGRSVAELVAL
MAV_4645|M.avium_104 YFTYRDVLGFIYAQAKFQVLGKENSNDVAAGRRKALAFIEGRSVEQLVAL
MLBr_02424|M.leprae_Br4923 YFTYRDVLGFIYAQAKFQLLGKENSQDVAAGQRKALAFIEGRSVEQLVAL
Mflv_0069|M.gilvum_PYR-GCK YFTYQDLARFAYAQAKFQLTGRENSDDVAAGRRKALSFIEGRSTAELVAL
Mvan_0843|M.vanbaalenii_PYR-1 YFTYQDLARFALAQAKFQLTGRENSGDVAAGRRKALAFIEGRSTAELVAL
MSMEG_0949|M.smegmatis_MC2_155 YFTYRDILGIAYAQAKFQLTGKENSDDVAAGRRKALAFIEGRSTAELVAL
TH_3424|M.thermoresistible__bu YFSYRDMLRFVYAQAKFQLTGRENSDDVAAGRRKALSFIEGRSVAELIAL
MAB_3995|M.abscessus_ATCC_1997 YFQYSDMLQFVWAQAKFRLTGRENSDDVAAGRQKALSFIEGRSVDELVAL
** * *: : *****:: * *** *****::***:******. :*: *
MMAR_0833|M.marinum_M GEEIYDEIIADKIWPGTRELAQMHLDAGQQVWLITATPYELAATIARRLG
MUL_4577|M.ulcerans_Agy99 GEEIYDDIIADKIWPGTRELAQMHLDAGQQVWLITATPYELAATIARRLG
Mb0517c|M.bovis_AF2122/97 GEEIYDEIIADKIWDGTRELTQMHLDAGQQVWLITATPYELAATIARRLG
Rv0505c|M.tuberculosis_H37Rv GEEIYDEIIADKIWDGTRELTQMHLDAGQQVWLITATPYELAATIARRLG
MAV_4645|M.avium_104 GEEIYDEIIADKIWPGTRELTQMHLDAGQQVWLITATPYELAATIARRLG
MLBr_02424|M.leprae_Br4923 GEEIYDEIIADKIWAGTRQLTQIHLDAGQQVWLITATPYELAATIARRLG
Mflv_0069|M.gilvum_PYR-GCK GEDIYDEIIADKIWPGTRRLAQMHLDAGQQVWLVTATPYELAETIARRLG
Mvan_0843|M.vanbaalenii_PYR-1 GEEIYDEIIADKIWPGTRALAQMHLDAGQQVWLVTATPYELADTIARRLG
MSMEG_0949|M.smegmatis_MC2_155 GEEIYDEIIADKIWPGTRALAQMHLDAGQQVWLVTATPYELAATIAKRLG
TH_3424|M.thermoresistible__bu GEEIYDEIIADKIWPGTLALAQMHLDAGQQVWLVTATPYELAATIARRLG
MAB_3995|M.abscessus_ATCC_1997 SEEIYDETIADKIWPGTRALTQMHLDAGQQVWLVTATPRELAETIARRLG
.*:***: ****** ** *:*:**********:**** *** ***:***
MMAR_0833|M.marinum_M LTGALGTVAESVDGVFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTA
MUL_4577|M.ulcerans_Agy99 LTGALGTVAESVDGVFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTA
Mb0517c|M.bovis_AF2122/97 LTGALGTVAESVDGIFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTA
Rv0505c|M.tuberculosis_H37Rv LTGALGTVAESVDGIFTGRLVGEILHGTGKAHAVRSLAIREGLNLKRCTA
MAV_4645|M.avium_104 LTGALGTVAESVDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLKRCTA
MLBr_02424|M.leprae_Br4923 LTGALGTVAESVDGIFTGRLVDELLHGVGKAHAVRSLAIREGLNLKRCTA
Mflv_0069|M.gilvum_PYR-GCK LTGALGTVAESIDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLRRCTA
Mvan_0843|M.vanbaalenii_PYR-1 LTGALGTVAESIDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLRRCTA
MSMEG_0949|M.smegmatis_MC2_155 FTGALGTVAESVDGVFTGRLVGDILHGTGKAHAVRSLAIREGLNLRRCTA
TH_3424|M.thermoresistible__bu FTGALGTVAESVDGVFTGRLVGDILHGVGKAHAVRNLAVREGLNLRRCTA
MAB_3995|M.abscessus_ATCC_1997 LTGALGTVAESVDGVFTGRLVGDILHGPGKARAVRNLAIRNGLNLKRCTA
:**********:**:******.::*** ***:***.**:*:****:****
MMAR_0833|M.marinum_M YSDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRTARKAARIG
MUL_4577|M.ulcerans_Agy99 YSDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRTARKAARIG
Mb0517c|M.bovis_AF2122/97 YSDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRIARKAARIG
Rv0505c|M.tuberculosis_H37Rv YSDSYNDVPMLSLVGTAVAINPDARLRSLARERGWEIRDFRIARKAARIG
MAV_4645|M.avium_104 YSDSYNDVPMLSLVGTAVAINPDARLRALARERGWEIRDFRTARKAARIG
MLBr_02424|M.leprae_Br4923 YSDSYNDVPMLSLVGTAVAINPDAQLRSLARERGWEIRDFRTARKAARIG
Mflv_0069|M.gilvum_PYR-GCK YSDSFNDVPMLSLVGTAVAINPDAALRDLARQRGWEVRDFRTARKAARIG
Mvan_0843|M.vanbaalenii_PYR-1 YSDSFNDVPMLSLVGTAVAINPDADLRDLARERGWEIRDFRTARKAARIG
MSMEG_0949|M.smegmatis_MC2_155 YSDSFNDVPMLSLVGTAVAINPDAALRDVARERGWEIRDFRTARKAARIG
TH_3424|M.thermoresistible__bu YSDSYNDVPMLSLVGTAVAINPDAALRSLARERGWEIRDFRTARKAARXX
MAB_3995|M.abscessus_ATCC_1997 YSDSVNDVPMLSLVGTAVAINPDAELRDVARRRGWEVRDFRTARKAARIG
**** ******************* ** :**.****:**** ******
MMAR_0833|M.marinum_M VPSALALGAAGGALAALASRRQSR--------------
MUL_4577|M.ulcerans_Agy99 VPSALALGAAGGALAALASRRQSR--------------
Mb0517c|M.bovis_AF2122/97 VPSALALGAAGGALAALASRRQSR--------------
Rv0505c|M.tuberculosis_H37Rv VPSALALGAAGGALAALASRRQSR--------------
MAV_4645|M.avium_104 VPSALALGAAGGALAALASRRQSR--------------
MLBr_02424|M.leprae_Br4923 VPSALALG---GALAAAVSRRRDRE-------------
Mflv_0069|M.gilvum_PYR-GCK VPSALALGAVGGALAAAVSHRDKK--------------
Mvan_0843|M.vanbaalenii_PYR-1 VPSALALGAVGGALAAAVSRREKK--------------
MSMEG_0949|M.smegmatis_MC2_155 VPSALALGALGGALAAVASRRHDLR-------------
TH_3424|M.thermoresistible__bu XSAARCSAAGRSASTRTWCRRWPTRSPRWCAAASRWRW
MAB_3995|M.abscessus_ATCC_1997 VPSAVLLGAAGGALAAALARREK---------------
.:* . .* : .:*