For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SIAANIIGTHYRYPDYFEVGREKVREFSAAVKDDHPAHFDEAAAKECGHDNLIAPLTFLAVAGRRVQLDL FDKFDVPINLERVLHRDQKLIFHRPIVVGDKLWFDSYLDSVIESHGTVICEIRAEVTDDDGKPVATSIVT MLGEAEIDEADEISSQIAAARDAAIAKMVAGQKSSS*
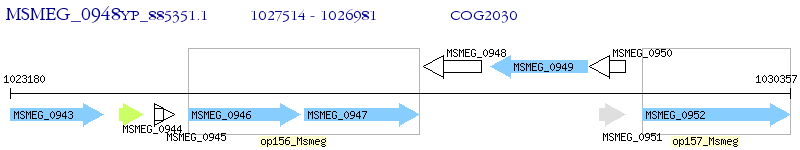
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0948 | - | - | 100% (177) | hypothetical protein MSMEG_0948 |
| M. smegmatis MC2 155 | MSMEG_1340 | - | 1e-31 | 41.78% (146) | (3R)-hydroxyacyl-ACP dehydratase subunit HadA |
| M. smegmatis MC2 155 | MSMEG_1342 | - | 1e-23 | 38.89% (144) | (3R)-hydroxyacyl-ACP dehydratase subunit HadC |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0516c | - | 2e-57 | 65.19% (158) | hypothetical protein Mb0516c |
| M. gilvum PYR-GCK | Mflv_0070 | - | 3e-73 | 74.29% (175) | MaoC-like dehydratase |
| M. tuberculosis H37Rv | Rv0504c | - | 2e-57 | 65.19% (158) | hypothetical protein Rv0504c |
| M. leprae Br4923 | MLBr_02425 | - | 2e-54 | 65.54% (148) | hypothetical protein MLBr_02425 |
| M. abscessus ATCC 19977 | MAB_3996 | - | 2e-52 | 63.10% (168) | hypothetical protein MAB_3996 |
| M. marinum M | MMAR_0832 | - | 1e-59 | 66.46% (158) | hypothetical protein MMAR_0832 |
| M. avium 104 | MAV_4646 | - | 2e-62 | 65.29% (170) | hypothetical protein MAV_4646 |
| M. thermoresistible (build 8) | TH_3830 | - | 2e-74 | 76.44% (174) | PUTATIVE MaoC-like dehydratase |
| M. ulcerans Agy99 | MUL_4576 | - | 7e-60 | 66.46% (158) | hypothetical protein MUL_4576 |
| M. vanbaalenii PYR-1 | Mvan_0842 | - | 5e-71 | 73.71% (175) | MaoC-like dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0832|M.marinum_M --MTVPPEAAGLIGKHYQAPDYFVVGREKIREFATAIQDEHPSHSDEAAA
MUL_4576|M.ulcerans_Agy99 --MTVPPEAAGLIGKHYQAPDYFVVGREKIREFATAIQDEHPSHSDEAAA
Mb0516c|M.bovis_AF2122/97 --MTVPEEAQTLIGKHYRAPDHFLVGREKIREFAVAVKDDHPTHYSEPDA
Rv0504c|M.tuberculosis_H37Rv --MTVPEEAQTLIGKHYRAPDHFLVGREKIREFAVAVKDDHPTHYSEPDA
MAV_4646|M.avium_104 --MTVPKEAEGLIGSHYRAPDYFEVGREKIREFALAVKDDHPAHFDESES
MLBr_02425|M.leprae_Br4923 --MTVPLEAEGLIGKHYRQLDHFQVGREKIREFAIAVKDDHPTHYNETAA
MSMEG_0948|M.smegmatis_MC2_155 --MSI---AANIIGTHYRYPDYFEVGREKVREFSAAVKDDHPAHFDEAAA
TH_3830|M.thermoresistible__bu --MSI---AADIIGTHYRYPDYFEVDREKIREFARAVKNDHPAHFTEEAA
Mflv_0070|M.gilvum_PYR-GCK MATSI---AADIIGTHYRYPDYFQVDREKIREFARAVKDEHPAHHDEDAA
Mvan_0842|M.vanbaalenii_PYR-1 --MAI---AEEIIGTHYRYPDYFAVDREKIREFARAVKDEHPAHYTEEAA
MAB_3996|M.abscessus_ATCC_1997 --MAI---RQDIVGTHYRYPDYFEVGREKMREFAAAIKDECEVN------
:: ::*.**: *:* *.***:***: *:::: :
MMAR_0832|M.marinum_M AELGYPELVAPLTFLAIAGRRVQLEIFNKFNIPINIARVFHRDQKFRFYR
MUL_4576|M.ulcerans_Agy99 TELGYPELVAPLTFLAIAGRRVQLEIFNKFNIPINIARVFHRDQKFRFYR
Mb0516c|M.bovis_AF2122/97 AAAGYPALVAPLTFLAIAGRRVQLEIFTKFNIPINIARVFHRDQKFRFHR
Rv0504c|M.tuberculosis_H37Rv AAAGYPALVAPLTFLAIAGRRVQLEIFTKFNIPINIARVFHRDQKFRFHR
MAV_4646|M.avium_104 AAAGYPDMVAPLTFLAIAGRRVQLEIFTKFSIPINIARVIHRDQKFKFHR
MLBr_02425|M.leprae_Br4923 FEAGYPALVAPLTFLAIAGRRVQLEIFTKFNIPINVARVFHRDQKFRFYR
MSMEG_0948|M.smegmatis_MC2_155 KECGHDNLIAPLTFLAVAGRRVQLDLFDKFDVPINLERVLHRDQKLIFHR
TH_3830|M.thermoresistible__bu RECGYDALIAPLTFLAVAGRRVQLEIFNHFDVPINLERMMHRDQKIIFHR
Mflv_0070|M.gilvum_PYR-GCK RECGHDALIASLTFLAVAGRRVQQEIFNQFDLPINMERVLHRDQKLVFHR
Mvan_0842|M.vanbaalenii_PYR-1 KEYGYDALVASLTFLAVAGRRVQEEIFNQFDLPINMERVLHRDQKLIFHR
MAB_3996|M.abscessus_ATCC_1997 -----SGVVAPITFLAVAGRRVQHKIFTEMDLPINVARVLHRDQKLKFHR
::*.:****:****** .:* .:.:***: *::*****: *:*
MMAR_0832|M.marinum_M PIVTGDKLYFHTYLDSVLESHGTVIAEIRSEVTDDEGKPVVTSVVTMLGE
MUL_4576|M.ulcerans_Agy99 PIVTGDKLYFHTYLDSVLESHGTVIAEIRSEVTDDEGKPVVTSVVTMLGE
Mb0516c|M.bovis_AF2122/97 PILANDKLYFDTYLDSVIESHGTVLAEIRSEVTDAEGKPVVTSVVTMLGE
Rv0504c|M.tuberculosis_H37Rv PILANDKLYFDTYLDSVIESHGTVLAEIRSEVTDAEGKPVVTSVVTMLGE
MAV_4646|M.avium_104 PILAHDRLYFDTYLDSVIESHGTVLAEIRSEVTDADGKPIVTSVVTMLGE
MLBr_02425|M.leprae_Br4923 TILAQDKLYFDTYLDSVIESHGTVIAEVRSEVTDTEGKAVVTSIVTMLGE
MSMEG_0948|M.smegmatis_MC2_155 PIVVGDKLWFDSYLDSVIESHGTVICEIRAEVTDDDGKPVATSIVTMLGE
TH_3830|M.thermoresistible__bu PIVAGDKLYFDSYLDSVIESHGTVITEVRGECTDADGNPVITSVVTILGE
Mflv_0070|M.gilvum_PYR-GCK PILAGDKLWFDSYLDSVTESHGAVLTEIRGEITDDEGNPIITSIVTVMGE
Mvan_0842|M.vanbaalenii_PYR-1 PILAGDKLWFDSYLDSVTESHGAVLTEIRGEVTDDNGEPVITSIVTVMGE
MAB_3996|M.abscessus_ATCC_1997 PIRVGDRLYFDSYLDSVLESHGMVTAEVRAEVTDENGDPVATSIVTMIGE
.* . *:*:*.:***** **** * *:*.* ** :*..: **:**::**
MMAR_0832|M.marinum_M AAH------------HEAEADAAVAAIASI-------------------
MUL_4576|M.ulcerans_Agy99 AAH------------HEAEADAAVAAIASI-------------------
Mb0516c|M.bovis_AF2122/97 AAH------------HEADADATVAAIASI-------------------
Rv0504c|M.tuberculosis_H37Rv AAH------------HEADADATVAAIASI-------------------
MAV_4646|M.avium_104 AAH------------QEADAAATAAAVASISAGKLGAI-----------
MLBr_02425|M.leprae_Br4923 LAR------------QDATAEETVAAIASI-------------------
MSMEG_0948|M.smegmatis_MC2_155 AEI-DEADEISSQIAAARDA-AIAKMVAGQKSSS---------------
TH_3830|M.thermoresistible__bu AAHPDEADELTAQIAARRDA-ALKQFVASQNRPATGGASAEEGAAPAGQ
Mflv_0070|M.gilvum_PYR-GCK AAHETEADELTAQIAAKRDADALAKFVAGQKGQID--------------
Mvan_0842|M.vanbaalenii_PYR-1 AESDTEADELTAQIAAKRDAAALSKFVAGQSPQAD--------------
MAB_3996|M.abscessus_ATCC_1997 AEG------------HERDAAAMAANIASHRKN----------------
* :*.