For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
AQQTPPATVKTDGRKRRWHQHKVERRNELVDGTLEAIRRRGSNVSMDEIAAEIGVSKTVLYRYFVDKNDL TTAVMMRFAQTTLIPNMAAALSSNLDGFALTREIIRVYVETVAAEPEPYRFVMANNSASKSKAVADSEQI IARMLAVMLRRRMAEVGMDTGGVEAWAFHTVGGVQLATHSWMSNPRMTSDELIDYLTMLSWYALCGIVEV GGSLEKFNSQPHPSPIPPRHLLD*
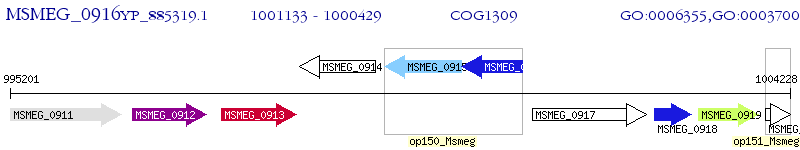
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0916 | - | - | 100% (234) | transcriptional regulator, TetR family protein |
| M. smegmatis MC2 155 | MSMEG_2309 | - | 8e-22 | 31.48% (216) | transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_1029 | - | 8e-22 | 31.48% (216) | transcriptional regulatory protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0482c | - | 1e-110 | 85.15% (229) | TetR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0103 | - | 1e-115 | 87.18% (234) | TetR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv0472c | - | 1e-110 | 85.15% (229) | TetR family transcriptional regulator |
| M. leprae Br4923 | MLBr_02457 | - | 1e-110 | 83.55% (231) | putative TetR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_4086 | - | 1e-101 | 78.60% (229) | TetR family transcriptional regulator |
| M. marinum M | MMAR_0797 | - | 1e-111 | 87.01% (231) | TetR family transcriptional regulator |
| M. avium 104 | MAV_4678 | - | 1e-102 | 83.56% (219) | transcriptional regulator, TetR family protein |
| M. thermoresistible (build 8) | TH_0865 | - | 1e-112 | 89.78% (225) | PROBABLE TRANSCRIPTIONAL REGULATORY PROTEIN (POSSIBLY |
| M. ulcerans Agy99 | MUL_4540 | - | 1e-110 | 88.79% (223) | TetR family transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_0805 | - | 1e-111 | 88.39% (224) | TetR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0103|M.gilvum_PYR-GCK ----MAQHTSPVAVKTDGRKRRWHQHKVERRNELVDGTLDAIRRLGSNVS
Mvan_0805|M.vanbaalenii_PYR-1 -----------MAAKTDGRKRRWHRHKVERRNELVDGTLEAIRRLGSNVS
MSMEG_0916|M.smegmatis_MC2_155 ----MAQQTPPATVKTDGRKRRWHQHKVERRNELVDGTLEAIRRRGSNVS
TH_0865|M.thermoresistible__bu ----VGQQTPPAAVKMDGRKRRWHQHKVERRNELVDGTLEAIRRRGSNVS
MMAR_0797|M.marinum_M ----MVERIPAMTGKTDGRKRRWHQHKVERRNELVDGTIVAIRRHGRFLS
MUL_4540|M.ulcerans_Agy99 -----------MTGKTDGRKRRWHQHKVERRNELVDGTIVAIRRHGRFLS
Mb0482c|M.bovis_AF2122/97 ----MAERIPAVTVKTDGRKRRWHQHKVERRNELVDGTIEAIRRHGRFLS
Rv0472c|M.tuberculosis_H37Rv ----MAERIPAVTVKTDGRKRRWHQHKVERRNELVDGTIEAIRRHGRFLS
MLBr_02457|M.leprae_Br4923 ----MAEHIPAVAVKTDGRKRRWHQHKVERRNELVDGTIDAIRRQGRFLS
MAV_4678|M.avium_104 ---------------MDGRKRRWHQHKVDRRNELVDGTIDAIRRLGGALS
MAB_4086|M.abscessus_ATCC_1997 MAITVASKTQPAAIKTDGRKRRWHKHKVERRTELVDGTLDAIRHRGRDIS
********:***:**.******: ***: * :*
Mflv_0103|M.gilvum_PYR-GCK MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFAQTTLIPNMAAALTSNL
Mvan_0805|M.vanbaalenii_PYR-1 MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFAQTTLIPNMAAALSSNL
MSMEG_0916|M.smegmatis_MC2_155 MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFAQTTLIPNMAAALSSNL
TH_0865|M.thermoresistible__bu MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFAQTTLIPNMAAALSTDL
MMAR_0797|M.marinum_M MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFAQTTLIPNMAAALSSNL
MUL_4540|M.ulcerans_Agy99 MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFAQTTLIPNMAAALSSNL
Mb0482c|M.bovis_AF2122/97 MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFTQTTLIPNMIAALSADM
Rv0472c|M.tuberculosis_H37Rv MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFTQTTLIPNMIAALSADM
MLBr_02457|M.leprae_Br4923 MDEIAGKIGVSKTVLYRYFIDKNDLTTAVMMRFAQTTLIPNMAAALSTNL
MAV_4678|M.avium_104 MDEIAAEIGVSKTVLYRYFVDKNDLTTAVMMRFTQTTLIPNMAAALTSGL
MAB_4086|M.abscessus_ATCC_1997 MDEIASDIGVSKTVLYRYFVDKNDLTTAVMTRFAQTTLIPKMAAALSARL
*****..************:********** **:******:* ***:: :
Mflv_0103|M.gilvum_PYR-GCK DGYELTREVIRVYVETVANEPEPYQFVMANNSASKSKAVADSEAIIARML
Mvan_0805|M.vanbaalenii_PYR-1 DGYDLTREIIKVYVETVAAEPEPYRFVMANNSASKSKAVADSETIIARML
MSMEG_0916|M.smegmatis_MC2_155 DGFALTREIIRVYVETVAAEPEPYRFVMANNSASKSKAVADSEQIIARML
TH_0865|M.thermoresistible__bu DGYDLTREIIRVYVETVAAEPEPYRFVMANNSAGKSKVVADSERIIARML
MMAR_0797|M.marinum_M DGFDLAREIIRVYVETVAAEPEPYRFVMANSSASKSKVIADSERIIARML
MUL_4540|M.ulcerans_Agy99 DGFDLAREIIRVYVETVAAEPEPYRFVMANSSASKSKVIADSERIIARML
Mb0482c|M.bovis_AF2122/97 DGFELTREIIRVYVETVAAQPEPYRFVMANSSASKSKVIADSERIIARML
Rv0472c|M.tuberculosis_H37Rv DGFELTREIIRVYVETVAAQPEPYRFVMANSSASKSKVIADSERIIARML
MLBr_02457|M.leprae_Br4923 DGFDLTREIIRVYVKTVAAEPEPYQFVMANSSASKSKVIADSERIIARML
MAV_4678|M.avium_104 DGFDLTREVIRVYVETVANEPEPYRFVMSNSSASKSKVIADSERIIARML
MAB_4086|M.abscessus_ATCC_1997 DGYDLTRTVIKVYVETVAAEPEIYPFVMANSSASRSKAIADSERIIARML
**: *:* :*:***:*** :** * ***:*.**.:**.:**** ******
Mflv_0103|M.gilvum_PYR-GCK AVMLRRRMAAAGMDASGAEPWAFHTVGGVQLATHSWMSNPRMTSDDLIDY
Mvan_0805|M.vanbaalenii_PYR-1 AVMLRRRMVSAGMDTGGAEPWAYHIVGGVQLATHSWMSNPRMTAEELIDY
MSMEG_0916|M.smegmatis_MC2_155 AVMLRRRMAEVGMDTGGVEAWAFHTVGGVQLATHSWMSNPRMTSDELIDY
TH_0865|M.thermoresistible__bu GVMLRRRMAEAGMDTRGVEPWSNLIVGGVQLATHSWMSHPRMTSDELIDY
MMAR_0797|M.marinum_M AVMLRRRMAEAGMDTGGVEPWAYLIVGGVQLATHSWMSDPRMTSDELIDY
MUL_4540|M.ulcerans_Agy99 AVMLRRRVAEAGMDTGGVEPWAYLIVGGVQLATHSWMSDPRMTSDELIDY
Mb0482c|M.bovis_AF2122/97 AVMLRRRMQEAGMDTGGVEPWAYLIVGGVQLATHSWMSDPRMSSDELIDY
Rv0472c|M.tuberculosis_H37Rv AVMLRRRMQEAGMDTGGVEPWAYLIVGGVQLATHSWMSDPRMSSDELIDY
MLBr_02457|M.leprae_Br4923 AVMIRRRMQEAGMDAGGVEPWAYLIVGGVQLATHSWMSDPRMSADELIDY
MAV_4678|M.avium_104 AVLMRRRMQHAGMDTGGVEPWAYLIVGGVQLATHSWMSYPRMSRDELIDY
MAB_4086|M.abscessus_ATCC_1997 ALMLRRRMHEAGMDTHGVEPWAYMIVGGVQLATHSWMSHPRMSSDELIDY
.:::***: .***: *.*.*: ************* ***: ::****
Mflv_0103|M.gilvum_PYR-GCK LTMLSWSALCGIVEVNGSLETFRQNPHPS---------------------
Mvan_0805|M.vanbaalenii_PYR-1 LTMLSWSALCGIVEVNGSLDTFRQSPHPS---------------------
MSMEG_0916|M.smegmatis_MC2_155 LTMLSWYALCGIVEVGGSLEKFNSQPHPS---------------------
TH_0865|M.thermoresistible__bu LTMLSWSALCGIVEVGGSLEKFREMPHPSXXSSSLSGNGLNSPQPWGPSI
MMAR_0797|M.marinum_M LTMLSWSALCGIVEAGGSLEKFRDQPHPS---------------------
MUL_4540|M.ulcerans_Agy99 LTMLSWSALCGIVEAGGSLEKFRDQPHPS---------------------
Mb0482c|M.bovis_AF2122/97 LTMLSWSALCGIVEAGGSLEKFREQPHPS---------------------
Rv0472c|M.tuberculosis_H37Rv LTMLSWSALCGIVEAGGSLEKFREQPHPS---------------------
MLBr_02457|M.leprae_Br4923 LTMLSWNALCGIVEVGGSLEKFRKQPHPS---------------------
MAV_4678|M.avium_104 LTMLSWNALKGIVEVGGSLEKFREEPHPS---------------------
MAB_4086|M.abscessus_ATCC_1997 LTMIAWNALLGIVDVGGSREKFVDMVHPS---------------------
***::* ** ***:..** :.* . ***
Mflv_0103|M.gilvum_PYR-GCK ---------PVLP-------------------SSLLD-------------
Mvan_0805|M.vanbaalenii_PYR-1 ---------PALP-------------------TRLLD-------------
MSMEG_0916|M.smegmatis_MC2_155 ---------PIPP-------------------RHLLD-------------
TH_0865|M.thermoresistible__bu SMPSRTAAPPDIPTVGIANSGAVSKSISVCTERKLADRARKRPEPGIRPG
MMAR_0797|M.marinum_M ---------PIVP-------------------PRVDD-------------
MUL_4540|M.ulcerans_Agy99 ---------PIVP-------------------PRVDD-------------
Mb0482c|M.bovis_AF2122/97 ---------PIVP-------------------AWGQV-------------
Rv0472c|M.tuberculosis_H37Rv ---------PIVP-------------------AWGQV-------------
MLBr_02457|M.leprae_Br4923 ---------PIIP-------------------PRGP--------------
MAV_4678|M.avium_104 ---------PIVP-------------------PREAAL------------
MAB_4086|M.abscessus_ATCC_1997 ---------PIPPP----------------PQTRQTHSA-----------
* *
Mflv_0103|M.gilvum_PYR-GCK -------
Mvan_0805|M.vanbaalenii_PYR-1 -------
MSMEG_0916|M.smegmatis_MC2_155 -------
TH_0865|M.thermoresistible__bu TRNRRRS
MMAR_0797|M.marinum_M -------
MUL_4540|M.ulcerans_Agy99 -------
Mb0482c|M.bovis_AF2122/97 -------
Rv0472c|M.tuberculosis_H37Rv -------
MLBr_02457|M.leprae_Br4923 -------
MAV_4678|M.avium_104 -------
MAB_4086|M.abscessus_ATCC_1997 -------