For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTVLVVNSGSSSLKYAVVRPASGEFLADGIIEEIGSGAVPDHDAALRAAFDELAAAGLHLEDLDLKAVGH RMVHGGKTFYKPSVVDDELIAKARELSPLAPLHNPPAIKGIEVARKLLPDLPHIAVFDTAFFHDLPAPAS TYAIDRELAETWHIKRYGFHGTSHEYVSQQAAIFLDRPLESLNQIVLHLGNGASASAVAGGKAVDTSMGL TPMEGLVMGTRSGDIDPGVIMYLWRTAGMSVDDIESMLNRRSGVLGLGGASDFRKLRELIESGDEHAKLA YDVYIHRLRKYIGAYMAVLGRTDVISFTAGVGENVPPVRRDALAGLGGLGIEIDDALNSAKSDEPRLIST PDSRVTVLVVPTNEELAIARACVGVV
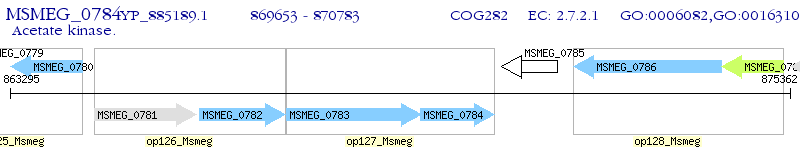
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0784 | ackA | - | 100% (376) | acetate kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0417 | ackA | 1e-142 | 66.31% (377) | acetate kinase |
| M. gilvum PYR-GCK | Mflv_0204 | - | 1e-152 | 70.48% (376) | acetate kinase |
| M. tuberculosis H37Rv | Rv0409 | ackA | 1e-142 | 66.31% (377) | acetate kinase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4225c | - | 1e-128 | 61.07% (375) | acetate kinase (AckA) |
| M. marinum M | MMAR_0711 | ackA | 1e-144 | 68.35% (376) | acetate kinase AckA |
| M. avium 104 | MAV_4758 | ackA | 1e-145 | 68.88% (376) | acetate kinase |
| M. thermoresistible (build 8) | TH_2306 | ackA | 1e-150 | 65.31% (392) | PROBABLE ACETATE KINASE ACKA (ACETOKINASE) |
| M. ulcerans Agy99 | MUL_2812 | ackA | 1e-144 | 67.82% (376) | acetate kinase |
| M. vanbaalenii PYR-1 | Mvan_0701 | - | 1e-152 | 70.71% (379) | acetate kinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0204|M.gilvum_PYR-GCK -----MTRTVLVLNSGSSSVKYAVLEPDSGVLIADGIVERIGQEG-----
Mvan_0701|M.vanbaalenii_PYR-1 -----MSRSVLVLNSGSSSVKYAVLEPDSGVLVADGIVERIGEEGAER--
MSMEG_0784|M.smegmatis_MC2_155 -------MTVLVVNSGSSSLKYAVVRPASGEFLADGIIEEIGSGA-----
Mb0417|M.bovis_AF2122/97 -----MSSTVLVINSGSSSLKFQLVEPVAGMSRAAGIVERIGERSS----
Rv0409|M.tuberculosis_H37Rv -----MSSTVLVINSGSSSLKFQLVEPVAGMSRAAGIVERIGERSS----
MMAR_0711|M.marinum_M MSASRPNRVVLVLNSGSSSLKFQLVEPDSGMSRATGNIERIGEESS----
MUL_2812|M.ulcerans_Agy99 ---------MLVLNSGSSSLKFQLVEPDSGMSRATGDIERIGEKSS----
MAV_4758|M.avium_104 MDGSDGARRVLVINSGSSSLKFQLVDPESGVAASTGIVERIGEESS----
TH_2306|M.thermoresistible__bu ----MSERNVLVINSGSSSLKFELIEADSGTSLAEGNVERIGEDTSLAGL
MAB_4225c|M.abscessus_ATCC_199 ---MSAGGSVLVLNCGSSSVKYQLIEPESGRVIAHGLVERIGEEE-----
:**:*.****:*: :: . :* : * :*.**.
Mflv_0204|M.gilvum_PYR-GCK ------------GAHDHAAAMRGVFDSLAADGHHLEDLGLVAVGHRVVHG
Mvan_0701|M.vanbaalenii_PYR-1 ------------AVTDHAAAMQVVFDSLASDGHRLDDLGLVAVGHRVVHG
MSMEG_0784|M.smegmatis_MC2_155 -------------VPDHDAALRAAFDELAAAGLHLEDLDLKAVGHRMVHG
Mb0417|M.bovis_AF2122/97 ------------PVADHAQALHRAFKMLAEDGIDLQTCGLVAVGHRVVHG
Rv0409|M.tuberculosis_H37Rv ------------PVADHAQALHRAFKMLAEDGIDLQTCGLVAVGHRVVHG
MMAR_0711|M.marinum_M ------------SVPDHDAALRRVFEILAEDDIDLQSCGLVAVGHRVVHG
MUL_2812|M.ulcerans_Agy99 ------------SVPDHDAALRRVFEILAEDDIDLQSCGLVAVGHRVVHG
MAV_4758|M.avium_104 ------------PVPDHDAALRRAFDMLAGDGVDLNTAGLVAVGHRVVHG
TH_2306|M.thermoresistible__bu RVGEREVRRDGEPIADHEAALRTAFDMFDEAGSNLAEMNLAAVGHRAVHG
MAB_4225c|M.abscessus_ATCC_199 -------------IGNHEEALRLVFDSIDTG-------DVVVVGHRVVHG
:* *:: .*. : .: .**** ***
Mflv_0204|M.gilvum_PYR-GCK GPDLYRPTIVDETVIDRLKELGPLAPLHNPPAVLGIEVARDALPDLPHVA
Mvan_0701|M.vanbaalenii_PYR-1 GQDLYRPTVVDDATIARLKELSPLAPLHNPPAILGIEVARKALPDLPHVA
MSMEG_0784|M.smegmatis_MC2_155 GKTFYKPSVVDDELIAKARELSPLAPLHNPPAIKGIEVARKLLPDLPHIA
Mb0417|M.bovis_AF2122/97 GTEFHQPTLLDDTVIGKLEELSALAPLHNPPAVLGIKVARRLLANVAHVA
Rv0409|M.tuberculosis_H37Rv GTEFHQPTLLDDTVIGKLEELSALAPLHNPPAVLGIKVARRLLANVAHVA
MMAR_0711|M.marinum_M GKDFYEPTLLNDAVIGKLDELSPLAPLHNPPAVLCIRVARALLPDVPHIA
MUL_2812|M.ulcerans_Agy99 GKDFYEPTLLNDAVIGKLDELSPLAPLHNPPAVLCLRVARALLPDVPHIA
MAV_4758|M.avium_104 GNTFYRPTVLDDAVIARLHELSELAPLHNPPALLGIEVARRLLPGIAHVA
TH_2306|M.thermoresistible__bu GQTFFEPTLVTDELIAKMAELAPLAPLHNPPGLLGMTVARKLLPDVPHVA
MAB_4225c|M.abscessus_ATCC_199 GQKFHEPTVLDDAVVAQIAELSSLAPLHNPAGVQGIEVARRLLPDVPQVA
* :..*::: : : : **. *******..: : *** *..:.::*
Mflv_0204|M.gilvum_PYR-GCK VFDTAFFHDLPAAAATYAIDAEVARDWNIRRYGFHGTSHQYVSEQAAAFL
Mvan_0701|M.vanbaalenii_PYR-1 VFDTAFFHDLPPAAATYAIDAEVAGSWHIRRYGFHGTSHQYVSEQAAAFL
MSMEG_0784|M.smegmatis_MC2_155 VFDTAFFHDLPAPASTYAIDRELAETWHIKRYGFHGTSHEYVSQQAAIFL
Mb0417|M.bovis_AF2122/97 VFDTAFFHDLPPAAATYAIDRDVADRWHIRRYGFHGTSHQYVSERAAAFL
Rv0409|M.tuberculosis_H37Rv VFDTAFFHDLPPAAATYAIDRDVADRWHIRRYGFHGTSHQYVSERAAAFL
MMAR_0711|M.marinum_M VFDTAFFHQLPPAAATYAIDRELADVWKIRRYGFHGTSHEYVSQQAAEFL
MUL_2812|M.ulcerans_Agy99 VFDTAFFHQLPPAAATYAIDRELADVWKIRRYGFHGTSHEYVSQQAADFL
MAV_4758|M.avium_104 VFDTGFFHDLPPAAATYAIDRELADRWQIRRYGFHGTSHRYVSEQAAAFL
TH_2306|M.thermoresistible__bu VFDTAFFHDLPAPAATYAIDREVAQRWDIRRYGFHGTSHQYVSEQAAVFL
MAB_4225c|M.abscessus_ATCC_199 VFDTAFFFGLPPAAAIYALDRAVAERYGIRRYGFHGTSHQYVSQQAAAFL
****.**. **..*: **:* :* : *:*********.***::** **
Mflv_0204|M.gilvum_PYR-GCK DVPLESLSQIVLHLGNGASVSAIVGGRPVETSMGLTPMEGLVMGTRSGDV
Mvan_0701|M.vanbaalenii_PYR-1 NVPLESLSQIVLHLGNGASASAILGGRPVETSMGLTPMEGLVMGTRSGDV
MSMEG_0784|M.smegmatis_MC2_155 DRPLESLNQIVLHLGNGASASAVAGGKAVDTSMGLTPMEGLVMGTRSGDI
Mb0417|M.bovis_AF2122/97 GRPLDGLNQIVLHLGNGASASAIARGRPVETSMGLTPLEGLVMGTRSGDL
Rv0409|M.tuberculosis_H37Rv GRPLDGLNQIVLHLGNGASASAIARGRPVETSMGLTPLEGLVMGTRSGDL
MMAR_0711|M.marinum_M GKPIGDLNQIVLHLGNGASASAVAGGRPVETSMGLTPLEGLVMGTRSGDL
MUL_2812|M.ulcerans_Agy99 SKPIGDLNQIVLHLGNGASVSAVAGGRPVETSMGLTPLEGLVMGTRSGDL
MAV_4758|M.avium_104 DRPLRGLKQIVLHLGNGCSASAIAGTRPLDTSMGLTPLEGLVMGTRSGDI
TH_2306|M.thermoresistible__bu DRPLSSLNQIVLHLGNGASVSAIVGGRAVDTSMGLTPLEGLVMGTRPGDI
MAB_4225c|M.abscessus_ATCC_199 GRGYADINQIVLHLGNGASASAILRGRAVETSMGLTPLEGLVMGTRTGDI
. .:.*********.*.**: :.::*******:********.**:
Mflv_0204|M.gilvum_PYR-GCK DPGVIFYLWREAGMPVEDIESMLNRRSGVRGLGGEIDFRELHRRIESGDD
Mvan_0701|M.vanbaalenii_PYR-1 DPGVIFYLWRTAGMSVEDIEAMLNRRSGVRGLGGEIDFRVLHQRIESGDA
MSMEG_0784|M.smegmatis_MC2_155 DPGVIMYLWRTAGMSVDDIESMLNRRSGVLGLGGASDFRKLRELIESGDE
Mb0417|M.bovis_AF2122/97 DPGVISYLWRTARMGVEDIESMLNHRSGMLGLAGERDFRRLRLVIETGDR
Rv0409|M.tuberculosis_H37Rv DPGVISYLWRTARMGVEDIESMLNHRSGMLGLAGERDFRRLRLVIETGDR
MMAR_0711|M.marinum_M DPGVIGYLWRTAKLGVDEIESMLNHRSGMLGLAGERDFRRLRAMIDDGDP
MUL_2812|M.ulcerans_Agy99 DPGVIGYLWRTAKLGVDEIESMLNHRSGMLALAGERDFRRLRAMIDDGDP
MAV_4758|M.avium_104 DPSVVSYLCHTAGMGVDDVESMLNHRSGVVGLSGVRDFRRLRELIESGDG
TH_2306|M.thermoresistible__bu DPGITFYLARTAGMTIDEIDELYNRRSGILGLAGANDFRELLKRIDAGDQ
MAB_4225c|M.abscessus_ATCC_199 DAGIVFHLARHG-MSIDEIDTVFNRRSGMLGLCGANDFREVHRLIDAGDA
*..: :* : . : ::::: : *:***: .* * *** : *: **
Mflv_0204|M.gilvum_PYR-GCK AAELAYEVYIHRLRKYIGAYLAILGSADVITFTAGVGENDAVVRRDALTG
Mvan_0701|M.vanbaalenii_PYR-1 AAQLAYDVYIHRLRKYVGAYLATLGSVDVITFTAGVGENDAAVRRDVLSG
MSMEG_0784|M.smegmatis_MC2_155 HAKLAYDVYIHRLRKYIGAYMAVLGRTDVISFTAGVGENVPPVRRDALAG
Mb0417|M.bovis_AF2122/97 SAQLAYEVFIHRLRKYLGAYLAVLGHTDVVSFTAGIGENDAAVRRDALAG
Rv0409|M.tuberculosis_H37Rv SAQLAYEVFIHRLRKYLGAYLAVLGHTDVVSFTAGIGENDAAVRRDALAG
MMAR_0711|M.marinum_M AAELAYDVFIHRLRKYVGAYLAVLGHTDVVSFTAGIGEHDAAVRRDTLAG
MUL_2812|M.ulcerans_Agy99 AAELAYDVFIHRLRKYVGAYLAVLGHTDVVSFTAGIGEHDAAVRRDALAG
MAV_4758|M.avium_104 AAQLAYSVFTHRLRKYIGAYLAVLGHTDVISFTAGIGENDAAVRRDAVSG
TH_2306|M.thermoresistible__bu AARLAYDIYIHRLRKYVGGYLALLGTTDVITFTAGVGENNPDVRRDALSG
MAB_4225c|M.abscessus_ATCC_199 AAQLAYNVYVHRLRKYIGAYIATLGGADVISFTAGAGENDALLRADALAG
*.***.:: ******:*.*:* ** .**::**** **: . :* *.::*
Mflv_0204|M.gilvum_PYR-GCK LAAFGIEIDEHLNDSPGRGARRISADGAPTTVLVIPTDEELAIARACTGV
Mvan_0701|M.vanbaalenii_PYR-1 LTAFGIELDEHLNASPARTARRISPDGAPITVLVVPTNEELAIARACAGV
MSMEG_0784|M.smegmatis_MC2_155 LGGLGIEIDDALNSAKSDEPRLISTPDSRVTVLVVPTNEELAIARACVGV
Mb0417|M.bovis_AF2122/97 LQGLGIALDQDRNLGPGHGARRISSDDSPIAVLVVPTNEELAIARDCLRV
Rv0409|M.tuberculosis_H37Rv LQGLGIALDQDRNLGPGHGARRISSDDSPIAVLVVPTNEELAIARDCLRV
MMAR_0711|M.marinum_M MAELGISLDERRNACPSGGARRISADDSPVTVLVIPTNEELAIARHCCSV
MUL_2812|M.ulcerans_Agy99 MAELGISLDEGRNACASGGARRISADDSPVTVLVIPTNEELAIARHCCSV
MAV_4758|M.avium_104 MEELGIVLDERRNLPGAKGARQISADDSPITVLVVPTNEELAIARDCVRV
TH_2306|M.thermoresistible__bu LGALGIELDEQLNAGPTRGPRRISAAGSPTTVLVIPTNEELAIARACLSV
MAB_4225c|M.abscessus_ATCC_199 LEVFGIEIDAARNQVRSGEARRISTDSSRVTVLVVPTNEELAIARAAVEA
: :** :* * .* **. .: :***:**:******* . .
Mflv_0204|M.gilvum_PYR-GCK LGARPGDG
Mvan_0701|M.vanbaalenii_PYR-1 LGGRLGHG
MSMEG_0784|M.smegmatis_MC2_155 V-------
Mb0417|M.bovis_AF2122/97 LGGRRA--
Rv0409|M.tuberculosis_H37Rv LGGRRA--
MMAR_0711|M.marinum_M LVAV----
MUL_2812|M.ulcerans_Agy99 LAAV----
MAV_4758|M.avium_104 LGG-----
TH_2306|M.thermoresistible__bu I-------
MAB_4225c|M.abscessus_ATCC_199 I-------
: