For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MGIETVVMSEPGRTHVQFVAVGQDDPLAAPLLDELADEYSSRYGSTRDAVMTWLLSQPAEVFAPPTGGMF VGLLAGEPVTGGAFQRFDDDTAELKRIWTHSAYRKRGLAKALLTHLEDQIAARGYRKVYLTTGHLQPEAE ALYAAAGYTRLPEPLPADGTVFPVAFHKVLPAEESA
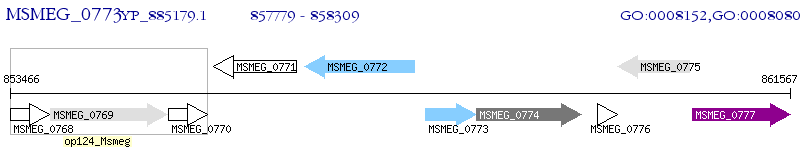
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0773 | - | - | 100% (176) | acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_0469 | - | 1e-10 | 30.50% (141) | transcriptional regulatory protein PadR- |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4235c | - | 9e-41 | 59.72% (144) | putative acetyltransferase, GNAT |
| M. marinum M | MMAR_0700 | - | 7e-53 | 62.50% (160) | hypothetical protein MMAR_0700 |
| M. avium 104 | MAV_4763 | - | 4e-46 | 55.09% (167) | acetyltransferase, gnat family protein |
| M. thermoresistible (build 8) | TH_2302 | - | 1e-50 | 63.23% (155) | acetyltransferase |
| M. ulcerans Agy99 | MUL_2823 | - | 5e-53 | 62.50% (160) | hypothetical protein MUL_2823 |
| M. vanbaalenii PYR-1 | Mvan_0695 | - | 4e-57 | 62.50% (168) | GCN5-related N-acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
TH_2302|M.thermoresistible__bu ---------------------VSTVEQLSFQAVGRHHRLAEPLLAELAVE
Mvan_0695|M.vanbaalenii_PYR-1 MTTPRGKLRRARPTLDSVVMSVQTVGELRFVAVGQDDPLAAPLIDELAVE
MMAR_0700|M.marinum_M ---------------MPEATQPTGAVELRFVPVALGDPLAQPLLTELAAE
MUL_2823|M.ulcerans_Agy99 ---------------MPEATQPTGAVELRFVPVALGDPLAQPLLTELAAE
MAV_4763|M.avium_104 ---------------MTPESSPPADGRLRFVPATHDDPLARPLLAELAVE
MSMEG_0773|M.smegmatis_MC2_155 ------------MGIETVVMSEPGRTHVQFVAVGQDDPLAAPLLDELADE
MAB_4235c|M.abscessus_ATCC_199 --------------------MTACEDLLQFVTVGQQDPLAAPLLAELAVE
: * .. . ** **: *** *
TH_2302|M.thermoresistible__bu YAHRYDSTVDRVARWLRSAPAAEFEPPDGALVIGLLSGQPVTGGAFRRFD
Mvan_0695|M.vanbaalenii_PYR-1 YADRYGGSRDKVHAWLRGYPAAEFEAPGGGLLIGLLDGQPVTGGAFRRFD
MMAR_0700|M.marinum_M YAQRYGVEQPAVLAWLQDGAALEFAPPDGGMLIGLRDGRPVTGGAFCRHD
MUL_2823|M.ulcerans_Agy99 YAQRYGVEQPAVLAWLQDGAALEFAPPDGGMLIGLRDGRPVTGGAFCRHD
MAV_4763|M.avium_104 YAQRYGGTPSAHLSWLPVPRD-ELAPPDGGLLIGVVDGVPVTGGAFRRYD
MSMEG_0773|M.smegmatis_MC2_155 YSSRYGSTRDAVMTWLLSQPAEVFAPPTGGMFVGLLAGEPVTGGAFQRFD
MAB_4235c|M.abscessus_ATCC_199 YSTRYGGTLAEHHDLLVNYPAAHFSAPDGGLLVGLQGGAAVTGGAFQRYD
*: **. * : .* *.:.:*: * .****** *.*
TH_2302|M.thermoresistible__bu AHTAELKRIWTDSRHRRRGHARALLAELERRITARGYRRIYLTTGDRQPE
Mvan_0695|M.vanbaalenii_PYR-1 DTTAELKRIWTDARYRRRGLGQALVSRLEADIAARGYSRIYLTTGDRQPE
MMAR_0700|M.marinum_M PDTAELKRIWTHREHRRRGHARALLARLEGEIAARGYRKVYLVTGNRQPE
MUL_2823|M.ulcerans_Agy99 PDTAELKRIWTHREHRRRGHARALLARLEGEIAARGYRKVYLVTGNRQPE
MAV_4763|M.avium_104 AETAELKRIWTDAAHRRRGYARALLAALEERTRACGYRRLYLITGNRQPE
MSMEG_0773|M.smegmatis_MC2_155 DDTAELKRIWTHSAYRKRGLAKALLTHLEDQIAARGYRKVYLTTGHLQPE
MAB_4235c|M.abscessus_ATCC_199 ANTAELKRIWTSSAHRRQGLAGRTLAALEVEIRRRGYRRIYLTTGPRQPE
********* :*::* . :: ** ** ::** ** ***
TH_2302|M.thermoresistible__bu AEALYRAAGYSRLP-KPLPGDEEGYPVAFLKELGGPGS-
Mvan_0695|M.vanbaalenii_PYR-1 AEALYSSLGYQRLE-EPLPAEGEVYAIAFLKELH-----
MMAR_0700|M.marinum_M AEELYRACGYTRLP-APQPSEGPVLPIAFGKALP-----
MUL_2823|M.ulcerans_Agy99 AEELYRACGYTRLP-APQPSEGPVLPIAFGKALP-----
MAV_4763|M.avium_104 AEALYDATGYTRVPPDPLPDWGPFRPIAFEKWLVPNER-
MSMEG_0773|M.smegmatis_MC2_155 AEALYAAAGYTRLP-EPLPADGTVFPVAFHKVLPAEESA
MAB_4235c|M.abscessus_ATCC_199 AKALYLSAGYRPLYDQTLTAD-EVGIHPFDKDL------
*: ** : ** : . . .* * *