For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
QPKPTVDTPIGELMSQLSAQTSRLVRDEMRLAQKEFTESAKHAGIGAGLFSAAGLIAVFGLAGVVTAAIA ALALVLPVWAAALIVAVVLFAIAGIIALISKKQIDQASPTPEMTVANVKQDIHEVQDEVRTGRHGQ*
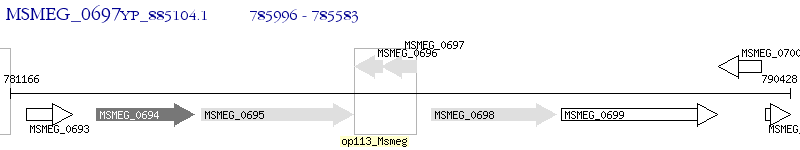
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0697 | - | - | 100% (137) | integral membrane protein |
| M. smegmatis MC2 155 | MSMEG_6182 | - | 2e-08 | 31.09% (119) | hypothetical protein MSMEG_6182 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3693 | - | 2e-06 | 25.23% (111) | transmembrane protein |
| M. gilvum PYR-GCK | Mflv_1378 | - | 5e-08 | 31.09% (119) | hypothetical protein Mflv_1378 |
| M. tuberculosis H37Rv | Rv3669 | - | 2e-06 | 25.23% (111) | transmembrane protein |
| M. leprae Br4923 | MLBr_02296 | - | 2e-06 | 27.97% (118) | hypothetical protein MLBr_02296 |
| M. abscessus ATCC 19977 | MAB_0423c | - | 2e-08 | 30.17% (116) | hypothetical protein MAB_0423c |
| M. marinum M | MMAR_5157 | - | 4e-06 | 28.83% (111) | hypothetical protein MMAR_5157 |
| M. avium 104 | MAV_0460 | - | 2e-06 | 29.66% (118) | hypothetical protein MAV_0460 |
| M. thermoresistible (build 8) | TH_4389 | - | 5e-46 | 67.65% (136) | PUTATIVE protein of unknown function DUF1469 |
| M. ulcerans Agy99 | MUL_4243 | - | 3e-06 | 28.83% (111) | hypothetical protein MUL_4243 |
| M. vanbaalenii PYR-1 | Mvan_0334 | - | 7e-38 | 61.90% (126) | hypothetical protein Mvan_0334 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5157|M.marinum_M MSKPDRK------NGVPSTLTTIPLADPHAM-PAEPSIGDLIKDATTQMS
MUL_4243|M.ulcerans_Agy99 MSKPDRK------NGVPSTLTTIPLADPHAM-PAEPSIGDLIKDATTQMS
MAV_0460|M.avium_104 MSKVEGK------NGVPSTLTTIPLTDPHAK-PAEPTIGDLIKDATTQVS
Mb3693|M.bovis_AF2122/97 MSKIDRK------NGVPSTLTTIPLADPHAG-PAEPSIGDLIKDATTQMS
Rv3669|M.tuberculosis_H37Rv MSKIDRK------NGVPSTLTTIPLADPHAG-PAEPSIGDLIKDATTQMS
MLBr_02296|M.leprae_Br4923 MSKGDRK------NGVPSTLTTIPLVDPHAE-PTEPSIGDLIKDATTQVS
Mflv_1378|M.gilvum_PYR-GCK MS--DRK------NDVPNTVTSIPLVDPHAP-KPDPSIGDLVKDATAQVS
MAB_0423c|M.abscessus_ATCC_199 MSSPGSRNYRTDSNGVPITVAAIPLSDPNATGTGEPTIGNLVRDATTQVS
MSMEG_0697|M.smegmatis_MC2_155 ---------------------------MQPKPTVDTPIGELMSQLSAQTS
TH_4389|M.thermoresistible__bu -------------------------MSLETKPAADASIGELLSQLSAQTS
Mvan_0334|M.vanbaalenii_PYR-1 ----------------------------MTHSKTEPSMGELVSQLSEQTS
. :..:*:*: : : * *
MMAR_5157|M.marinum_M TLVRAEVELARAEITRDVKKGLTGSVFFIAALVVLFYSTFFLFFFLAELL
MUL_4243|M.ulcerans_Agy99 TLVRAEVELARAEITRDVKKGLTGSVFFIAALVVLFYSTFFLFFFLAELL
MAV_0460|M.avium_104 TLVRAEVELARAEITRDVKKGLTGSVFFIAALVVLFYSTFFFFFFVAELL
Mb3693|M.bovis_AF2122/97 TLVRAEVELARAEITRDVKKGLTGSVFFISSLVVGFYSTFFFFFFVAELL
Rv3669|M.tuberculosis_H37Rv TLVRAEVELARAEITRDVKKGLTGSVFFISSLVVGFYSTFFFFFFVAELL
MLBr_02296|M.leprae_Br4923 TLVRAEVELARAEIIRDVKKGLTGSVFFIAALVVLFYSTFFFFLFLAELL
Mflv_1378|M.gilvum_PYR-GCK TLVRAEVELAKAEITRDVKKGLTGSVFFIAALVVLFYSTFFFFFFLAELL
MAB_0423c|M.abscessus_ATCC_199 ALVRAEVELARAEVIRDVKKGVAGSIFFIAALVVLFYSTFFFFFFAAEVL
MSMEG_0697|M.smegmatis_MC2_155 RLVRDEMRLAQKEFTESAKHAGIGAGLFSAAGLIAVFGLAGVVTAAIAAL
TH_4389|M.thermoresistible__bu RLVRDELRLAQQEFRESARHAGIGAGLFSAAGVLALFGVAALLTAAVAAL
Mvan_0334|M.vanbaalenii_PYR-1 RLVRDEMRLAQKEMQESAKHAGLGAGMVGAAALLSVLGLATLIAGAVAAL
*** *:.**: *. ...::. *: :. :: :: . . .. *
MMAR_5157|M.marinum_M DTWLWRWVAFLIVFGLMVLTTGMLALLGFLKVRRIR-GPRQTIESVKETR
MUL_4243|M.ulcerans_Agy99 DTWLWRWVAFLIVFGLMVLTTGMLALLGFLKVRRIR-GPRQTIESVKETR
MAV_0460|M.avium_104 DTWLWRWVAFLIVFGVQVVVGAVLALLGFLKVRRIR-GPRQTIESVKETR
Mb3693|M.bovis_AF2122/97 DTWIWRWVAFLLVFAIMVVVTAVLALLGFLKVRRIR-GPRQTIASVKETR
Rv3669|M.tuberculosis_H37Rv DTWIWRWVAFLLVFAIMVVVTAVLALLGFLKVRRIR-GPRQTIASVKETR
MLBr_02296|M.leprae_Br4923 DTWLWRWVALLIVFAIMVMVTAALALLGYVKIRRIR-GPHQTIESVKETR
Mflv_1378|M.gilvum_PYR-GCK DNWLWKWAAYLIVFGIMVVLTALLALFGYLKVRRIR-GPQKTIETVKELP
MAB_0423c|M.abscessus_ATCC_199 KIWLPAWAAYLIIFAAMVFVAVLAVLIGYLQVRRIR-GPQATIESVKETA
MSMEG_0697|M.smegmatis_MC2_155 ALVLPVWAAALIVAVVLFAIAGIIALISKKQIDQASPTPEMTVANVKQDI
TH_4389|M.thermoresistible__bu ALVLPVWAAALIVAAALLLIAGIAALISKKQVDSVTPAAPRTVQTVKDDI
Mvan_0334|M.vanbaalenii_PYR-1 ALALPVWLSAVIVAVVVFAAAGIAALVGKKQAEQVPPPAAQSVDSVKSDI
: * : ::: . .*.. : . :: .**.
MMAR_5157|M.marinum_M TALIPGHDKST----AVEPSASAP-GKP----VTDPSGW
MUL_4243|M.ulcerans_Agy99 TALIPGHDKST----AVEPSAPAP-GKP----VTDPSGW
MAV_0460|M.avium_104 TALTPGHDKAAG-ARALTESHGKH-AKPGDGAAADPSGW
Mb3693|M.bovis_AF2122/97 TALTPGHDKTP---VTPKPVTSDR-ATP-----VDPSGW
Rv3669|M.tuberculosis_H37Rv TALTPGHDKTP---VTPKPVTSDR-ATP-----VDPSGW
MLBr_02296|M.leprae_Br4923 TALTPGHDKAQARPRKLTGSGTNPENNPDRRTPADPSGW
Mflv_1378|M.gilvum_PYR-GCK DAFTPGPDKRA-----VAASKDNLPATQ-----KDPSGW
MAB_0423c|M.abscessus_ATCC_199 NVFS-REDEAP---------SDTKAIKP-----VDPSGW
MSMEG_0697|M.smegmatis_MC2_155 HEVQDEVRTGR---------HGQ----------------
TH_4389|M.thermoresistible__bu R----EVKEAR---------HG-----------------
Mvan_0334|M.vanbaalenii_PYR-1 D----EIKVAR---------HGSRT--------------