For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTGPVNPDDRRSFSSRTPVNENPDGVQYRRGFVTRHQVSGWRFVMRRIASGVALHDTRMLVDPLRTQSRA VLTGALILVTGLVGCFIFSLFRPGGVPGNNAILADRSTSALYVRVGEQLHPVLNLTSARLISGSPDNPTM VKTSEIDKFPRGNLLGIPGAPERMVQNAATDAEWTVCDAVGGANPGVTVIAGPLGADGERAAPLPPDHAV LVHSDAEPNPGDWLLWDGKRSPIDLADRAVTDALGLGGQALAPRPIAAGLFNAVPAAPALTAPVIPDAGA APQFELSLPVPVGAVVVAYDADNTARYYAVLSDGLQPISPVLAAILRNTDSHGFAQPPRLGPDEVARTPM SRGLDTSAYPDNPVTLVEASAHPVTCAHWTKPSDAAESSLSVLSGAVLPLAEGLHTVDLVGAGAGGAANR VALTPGTGYFVQTVGAEPGSPTAGSMFWVSDTGVRYGIDTAEDDKVVAALGLSTSPLPVPWSVLSQFAAG PALSRGDALVAHDAVSTNPNSARMEASR
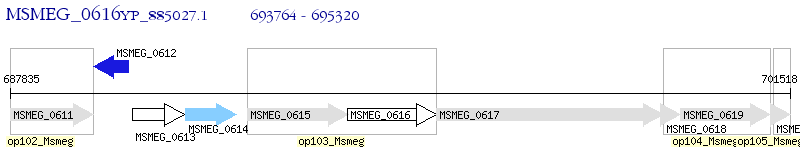
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0616 | - | - | 100% (518) | hypothetical protein MSMEG_0616 |
| M. smegmatis MC2 155 | MSMEG_0060 | - | 2e-70 | 35.10% (490) | hypothetical protein MSMEG_0060 |
| M. smegmatis MC2 155 | MSMEG_1534 | - | 2e-62 | 36.04% (480) | hypothetical protein MSMEG_1534 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0291 | - | 0.0 | 66.16% (523) | hypothetical protein Mb0291 |
| M. gilvum PYR-GCK | Mflv_0328 | - | 0.0 | 65.23% (512) | hypothetical protein Mflv_0328 |
| M. tuberculosis H37Rv | Rv0283 | - | 0.0 | 66.16% (523) | hypothetical protein Rv0283 |
| M. leprae Br4923 | MLBr_02536 | - | 0.0 | 61.54% (533) | hypothetical protein MLBr_02536 |
| M. abscessus ATCC 19977 | MAB_2233c | - | 1e-159 | 57.00% (507) | hypothetical protein MAB_2233c |
| M. marinum M | MMAR_0542 | - | 0.0 | 63.64% (517) | hypothetical protein MMAR_0542 |
| M. avium 104 | MAV_4870 | - | 0.0 | 64.88% (541) | hypothetical protein MAV_4870 |
| M. thermoresistible (build 8) | TH_2593 | - | 3e-84 | 65.94% (229) | PUTATIVE protein of unknown function DUF690 |
| M. ulcerans Agy99 | MUL_1205 | - | 0.0 | 63.25% (517) | hypothetical protein MUL_1205 |
| M. vanbaalenii PYR-1 | Mvan_0412 | - | 0.0 | 66.41% (512) | hypothetical protein Mvan_0412 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0542|M.marinum_M MTTNEPDGEWAGARRSFTSRTPVNDNPDKVEYRRGFVTRHQVTGWRFVMR
MUL_1205|M.ulcerans_Agy99 MTTNEPDGEWAGARRSFTSRTPVNDNPDKVEYRRGFVTRHQVTGWRFVMR
Mb0291|M.bovis_AF2122/97 MTNQQHDHDFDHDRRSFASRTPVNNNPDKVVYRRGFVTRHQVTGWRFVMR
Rv0283|M.tuberculosis_H37Rv MTNQQHDHDFDHDRRSFASRTPVNNNPDKVVYRRGFVTRHQVTGWRFVMR
MLBr_02536|M.leprae_Br4923 MTSNELPGEWSGERRSFFSRTPVNDNPDKVVYRRGFVTRHQVTGWRFVMR
MAV_4870|M.avium_104 MTANNDPD----DRRSFTSRTPVNDNPDQVQYRRGFVTRHQVSGWRFVMR
MAB_2233c|M.abscessus_ATCC_199 -MSDNRDNNANSERRAFASRTPASENPEPLKYRRGFVTGHQVTGWRFVMR
MSMEG_0616|M.smegmatis_MC2_155 ----MTGPVNPDDRRSFSSRTPVNENPDGVQYRRGFVTRHQVSGWRFVMR
TH_2593|M.thermoresistible__bu --------------------------------------------------
Mflv_0328|M.gilvum_PYR-GCK ------MSGADGDRRSFTSRTPTNENPDRVSFRRGFVTRHQVSGWRFMVR
Mvan_0412|M.vanbaalenii_PYR-1 ------MSAPEEERRSFASRTPTNGNPDRVTYRRGFVTRHQVTGWRFVMR
MMAR_0542|M.marinum_M RIASGIALHDTRMLVDPLRTQSRSVLMGVLIFITGLVGCFVFSLIRPNGQ
MUL_1205|M.ulcerans_Agy99 RIASGIALHDTRMLVDPLRTQSRSVLMGVLIFITGLVGCFVFSLIRPNGQ
Mb0291|M.bovis_AF2122/97 RIAAGIALHDTRMLVDPLRTQSRAVLMGVLIVITGLIGSFVFSLIRPNGQ
Rv0283|M.tuberculosis_H37Rv RIAAGIALHDTRMLVDPLRTQSRAVLMGVLIVITGLIGSFVFSLIRPNGQ
MLBr_02536|M.leprae_Br4923 RIASGIALHDTRMLVDPLRTQSRSVLVGALLVITGLIGCFVFSFIRPNGA
MAV_4870|M.avium_104 RIASGIALHDTRMLVDPLRTQSRAVAMGAVLLVTGLAGCFVFSLIRPNGT
MAB_2233c|M.abscessus_ATCC_199 RIASGVALHDARMLVDPLRTQSRAFGMGAVVVVTGLVGCFVFSLLRPSGI
MSMEG_0616|M.smegmatis_MC2_155 RIASGVALHDTRMLVDPLRTQSRAVLTGALILVTGLVGCFIFSLFRPGGV
TH_2593|M.thermoresistible__bu --------------------------------------------------
Mflv_0328|M.gilvum_PYR-GCK RMASGVALHDTRMLVDPLRTQSRSVLVGALILVTGLIGCFLFSLIRPAGD
Mvan_0412|M.vanbaalenii_PYR-1 RIASGVALHDTRMLVDPLRTQSRSVLVGALILVTGLIGCFLFSLIRPSGA
MMAR_0542|M.marinum_M VGSNAVLADRSTAALYVRVGDTLHPVLNLTSARLIVGHPVNPTTVKSTEL
MUL_1205|M.ulcerans_Agy99 VGSNAVLADRSTAALYVRVGDTLHPVLNLTSARLIVGHPVNPTTVKSTEL
Mb0291|M.bovis_AF2122/97 AGSNAVLADRSTAALYVRVGEQLHPVLNLTSARLIVGRPVSPTTVKSTEL
Rv0283|M.tuberculosis_H37Rv AGSNAVLADRSTAALYVRVGEQLHPVLNLTSARLIVGRPVSPTTVKSTEL
MLBr_02536|M.leprae_Br4923 AGNNAVLADRSTAALYVRVGDELHPVLNLTSARLIVGRSVNPITVKSSEL
MAV_4870|M.avium_104 AGTNAVLADRSTAALYVRVGDDLHPVLNLTSARLIVGKPVNPTTVKSAEL
MAB_2233c|M.abscessus_ATCC_199 AGTDPILADRSTAALYVRVGDNLHPVLNLTSARLIAGKAADPTMVKSTQL
MSMEG_0616|M.smegmatis_MC2_155 PGNNAILADRSTSALYVRVGEQLHPVLNLTSARLISGSPDNPTMVKTSEI
TH_2593|M.thermoresistible__bu --------------------------------------------------
Mflv_0328|M.gilvum_PYR-GCK VGTDSILAERDTAALYVRLGDQVHPVLNLASARLITGRPDNPAMVKSSEL
Mvan_0412|M.vanbaalenii_PYR-1 AGTDAVLADRDTAALYVRLGEQLHPVLNLTSARLIAGRPDNPAMVKSSEL
MMAR_0542|M.marinum_M DRFPRGNLIGIPGAPERMVQSEARDSDWTVCDAISSGSQGGSAAHSTGVT
MUL_1205|M.ulcerans_Agy99 DRFPRGNLIGIPGAPERMVQSEARDSDWTVCDAISSGSQGGSAAHSTGVT
Mb0291|M.bovis_AF2122/97 DQFPRGNLIGIPGAPERMVQNTSTDANWTVCDGLNAPSRGG--ADGVGVT
Rv0283|M.tuberculosis_H37Rv DQFPRGNLIGIPGAPERMVQNTSTDANWTVCDGLNAPSRGG--ADGVGVT
MLBr_02536|M.leprae_Br4923 DRFPRGNLIGIPGAPERMVQNTTHDANWTVCDVVSG--EGGHAAHSMGVT
MAV_4870|M.avium_104 DRFPRGNLLGIPGAPERMVQNPSTDANWTVCDSVSEPAPGGHGAHSTGVT
MAB_2233c|M.abscessus_ATCC_199 DKFPRGNLIGIPGAPERMVQAKATDAAWAVCEAGQTS------TDNRSVT
MSMEG_0616|M.smegmatis_MC2_155 DKFPRGNLLGIPGAPERMVQNAATDAEWTVCDAVGG--------ANPGVT
TH_2593|M.thermoresistible__bu --------------------------------------------------
Mflv_0328|M.gilvum_PYR-GCK DELPRGNMLGIPGAPGRMAVNTTRDAYWTVCDAAAG--------AVTGVT
Mvan_0412|M.vanbaalenii_PYR-1 DQFPRGNLIGIPGAPERMVANTTRDAYWTVCDAATG--------STAGVT
MMAR_0542|M.marinum_M VIAGSPEPSGHRATALGAGQVILVND----GTSSWLLWDGKRSRMDLADH
MUL_1205|M.ulcerans_Agy99 VIAGSPEPSGHRATALGAGQVILVND----GTSSWLLWDGKRSRMDLADH
Mb0291|M.bovis_AF2122/97 VIAGPLEDTGARAAALGPGQAVLVDS----GAGTWLLWDGKRSPIDLADH
Rv0283|M.tuberculosis_H37Rv VIAGPLEDTGARAAALGPGQAVLVDS----GAGTWLLWDGKRSPIDLADH
MLBr_02536|M.leprae_Br4923 VIAGPPDSHGMRAAVLGSAHGVLVDAPSERGGSTWLLWDGKRSEIDLADH
MAV_4870|M.avium_104 VIAGRPDSSGARAAALPSQQAILAER----DGGTWLLWNGRRSQINLADH
MAB_2233c|M.abscessus_ATCC_199 VIAGPLDTDGDKAADLPRDRAAVVSS----EAGTWLLWDGKRSALNLGDV
MSMEG_0616|M.smegmatis_MC2_155 VIAGPLGADGERAAPLPPDHAVLVHSDAEPNPGDWLLWDGKRSPIDLADR
TH_2593|M.thermoresistible__bu --------------------------------------------------
Mflv_0328|M.gilvum_PYR-GCK VIAGAPDDTGERATVLPDGHAVLAEN----NGRTWLLWDGRRSAIDLADR
Mvan_0412|M.vanbaalenii_PYR-1 VIAGRPADGGERAAALPAGHAVLAEN----GGRTWLLWDGKRSPIDLADR
MMAR_0542|M.marinum_M AVTNALGFGT---DLPAPRPIAPGLFNAIPEAAPLAAPAIPDAGNPAAFA
MUL_1205|M.ulcerans_Agy99 AVTNALGFAT---DLPAPRPIAPGLFNAIPEAAPLAAPAIPDAGNPAAFA
Mb0291|M.bovis_AF2122/97 AVTSGLGLGA---DVPAPRIIASGLFNAIPEAPPLTAPIIPDAGNPASFG
Rv0283|M.tuberculosis_H37Rv AVTSGLGLGA---DVPAPRIIASGLFNAIPEAPPLTAPIIPDAGNPASFG
MLBr_02536|M.leprae_Br4923 AVTDALGFGVGFAEVPAPRPIGAGLFNAIPEAPPLKAPVIPNAGATPSFG
MAV_4870|M.avium_104 AVTNALGLTENNSELPAPRPIALGLFNAIPEAPSLSAPGIPNAGAPAPFA
MAB_2233c|M.abscessus_ATCC_199 AVTSALGFTAP----PAVRPVTTRLLNAIPEAPALVIPAIAGAGAPTSYP
MSMEG_0616|M.smegmatis_MC2_155 AVTDALGLGGQ---ALAPRPIAAGLFNAVPAAPALTAPVIPDAGAAPQFE
TH_2593|M.thermoresistible__bu --------------------------------------------------
Mflv_0328|M.gilvum_PYR-GCK AVAGGVGIGVG---VAAPQPIATGLFNAIPEAPALITPVIPNAGLPSDLG
Mvan_0412|M.vanbaalenii_PYR-1 AVTGGLGIGVD---SAAPQPISVGLFNAIPEGPPLVTPAIPGAGSPSAAP
MMAR_0542|M.marinum_M VP--GPIGSVVQAYTLGD------NSIQYYAVLPDGLQPISPVLAAILRN
MUL_1205|M.ulcerans_Agy99 VP--GPIGSVVQAYTLGD------NSIQYYAVLPDGLQPISPVLAAIVRN
Mb0291|M.bovis_AF2122/97 VP--APIGAVVSSYALKDSGKTISDTVQYYAVLPDGLQQISPVLAAILRN
Rv0283|M.tuberculosis_H37Rv VP--APIGAVVSSYALKDSGKTISDTVQYYAVLPDGLQQISPVLAAILRN
MLBr_02536|M.leprae_Br4923 VR--APIGAVVVSFGLAALGKNPYDSVRYYAVLPDGLQPISPVLAAILRN
MAV_4870|M.avium_104 VP--APIGAVVVSYTLDQSS---SGAPRYYAVLPDGLQQISPVLAAILRN
MAB_2233c|M.abscessus_ATCC_199 LPVAAPIGSVLVSYSTDN-------TPVHYAVLTDGVQQISPVIASALRN
MSMEG_0616|M.smegmatis_MC2_155 LSLPVPVGAVVVAYDADN-------TARYYAVLSDGLQPISPVLAAILRN
TH_2593|M.thermoresistible__bu ------VGAVVTAYEADN-------TIRYYAVLPDGLQPVPVVLASIMRN
Mflv_0328|M.gilvum_PYR-GCK LPEPFPVGSVVVSYDEDN-------TLRHYVVLTDGLQPISPVVAAILRN
Mvan_0412|M.vanbaalenii_PYR-1 LPEPLPIGAVVVAYDADN-------AIRHYVVLADGLQAVSPVVAAILRN
:*:*: :: : :*.**.**:* :. *:*: :**
MMAR_0542|M.marinum_M TNSYGLQQPPRIGADDVAKLPVSRMLDTTRYPATPMHLVDTESNPITCAY
MUL_1205|M.ulcerans_Agy99 TNSYGLQQPPRIGADDVAKLPVSRMLDTTRYPATPMHLVDTESNPITCAY
Mb0291|M.bovis_AF2122/97 NNSYGLQQPPRLGADEVAKLPVSRVLDTRRYPSEPVSLVDVTRDPVTCAY
Rv0283|M.tuberculosis_H37Rv NNSYGLQQPPRLGADEVAKLPVSRVLDTRRYPSEPVSLVDVTRDPVTCAY
MLBr_02536|M.leprae_Br4923 INSYGLQQPPRLGADEVDKLPVSRMLDTERYPEQQISLIDAGYAPVSCAY
MAV_4870|M.avium_104 TNSYGLDQPPRLGADAVAKLPVSRMLDTARYPDQHVSLVDTAKSPVTCAY
MAB_2233c|M.abscessus_ATCC_199 ANSYGLEQPPRLTADQIAHLPTSSAIDTRVFPNVAVKPLAADEDPVLCTR
MSMEG_0616|M.smegmatis_MC2_155 TDSHGFAQPPRLGPDEVARTPMSRGLDTSAYPDNPVTLVEASAHPVTCAH
TH_2593|M.thermoresistible__bu TDSYGFAQPPRLGMDEIARLPVADTLDTDAYPARPVTLVNADAKPMTCVH
Mflv_0328|M.gilvum_PYR-GCK SNSWGLASPPRLDADLVARTPQAEVVDVDAYPAERVTLVDTAAEPVICAA
Mvan_0412|M.vanbaalenii_PYR-1 SNSYGLAQPPRLDADVVARIPEAAGVDNAAYPSDRVTLVDVAADPVTCAQ
:* *: .***: * : : * : :* :* : : . *: *.
MMAR_0542|M.marinum_M WSKPVGATTSSLTLLSGAALPVPDAVHTVDLVGGGSQ-NGAVATRVALAP
MUL_1205|M.ulcerans_Agy99 WSKPVGATTSSLTLLSGAALPVPGAVHTVDLVGGGSQ-NGAVATRVALAP
Mb0291|M.bovis_AF2122/97 WSKPVGAATSSLTLLAGSALPVPDAVHTVELVGAG---NGGVATRVALAA
Rv0283|M.tuberculosis_H37Rv WSKPVGAATSSLTLLAGSALPVPDAVHTVELVGAG---NGGVATRVALAA
MLBr_02536|M.leprae_Br4923 WSKPAGAATSFLSLMSGAALPVPDAARAVELVSAPSRGDSSTASRVVLTP
MAV_4870|M.avium_104 WSKPAGAATSSLNLLSGSALPIAESIRPVDLVGGGSPQNGPVATRVALPP
MAB_2233c|M.abscessus_ATCC_199 FTRTADATADSWSLLAGSVLPLSDSVHTVQLISPA-----ATPTRVAIAP
MSMEG_0616|M.smegmatis_MC2_155 WTKPSDAAESSLSVLSGAVLPLAEGLHTVDLVGAGA---GGAANRVALTP
TH_2593|M.thermoresistible__bu WSKPADAPANSLTLLSGAALPLPDGQHTVDLVSAGA---GGTADRVALPA
Mflv_0328|M.gilvum_PYR-GCK WSEHEGAQESSMELRSGAALPVADGVSTVALVSAGAP--GGPANQVALPQ
Mvan_0412|M.vanbaalenii_PYR-1 WAKPEGAQDASLTLRTGAVLPVADGARTVELVSSGTP--GGPADRVALPP
::. .* : :*:.**:. . .* *:. . :*.:.
MMAR_0542|M.marinum_M GTGYFTQTAGGGPAAPPTG--SLFWVSDTGVRYGIDTEGE-------GGR
MUL_1205|M.ulcerans_Agy99 GTGYFTQTAGGGPAAPPTG--SLFWVSDTGVRYGIDTEGE-------GGR
Mb0291|M.bovis_AF2122/97 GTGYFTQTVGGGPDAPGAG--SLFWVSDTGVRYGIDNEPQG----VAGGG
Rv0283|M.tuberculosis_H37Rv GTGYFTQTVGGGPDAPGAG--SLFWVSDTGVRYGIDNEPQG----VAGGG
MLBr_02536|M.leprae_Br4923 GTGYFAQTVGVGSAAPATA--SLFWVSDTGVRYGIDTEADARSEATAGPG
MAV_4870|M.avium_104 GTGYFTQTASGGAGSPGAG--SLFWVSDTGVRYGIDNEAEHS---AAGHG
MAB_2233c|M.abscessus_ATCC_199 GTGYLVQPGGRG---------GYQWISDTGVRYGIDTEAE--------GD
MSMEG_0616|M.smegmatis_MC2_155 GTGYFVQTVGAEPGSPTAG--SMFWVSDTGVRYGIDTAED---------D
TH_2593|M.thermoresistible__bu GTGYLVDSVGLRTGADSAEGPGLFWISDTGVRYGIDTHGD---------D
Mflv_0328|M.gilvum_PYR-GCK GTGYFVDNAGS-----------LFWLSDTGVRYGIDTGADA------DGA
Mvan_0412|M.vanbaalenii_PYR-1 GTGYFVDSAGS-----------LFWISDTGVRYGIDTGS--------DGV
****:.: . *:**********.
MMAR_0542|M.marinum_M KTVEALGLQDPPVLIPWSVLSLFAPGPTLSRADALMAHDGLAPDSKPGRP
MUL_1205|M.ulcerans_Agy99 KTVEALGLQDPPVLIPWSVLSLFAPGPTLSRADALMAHDGLAPDSKPGRP
Mb0291|M.bovis_AF2122/97 KAVEALGLNPPPVPIPWSVLSLFVPGPTLSRADALLAHDTLVPDSRPARP
Rv0283|M.tuberculosis_H37Rv KAVEALGLNPPPVPIPWSVLSLFVPGPTLSRADALLAHDTLVPDSRPARP
MLBr_02536|M.leprae_Br4923 KIVEALGLKLPAVPIPWSILSLFAVGPTLSRADALLEHDGLAPDTRAGRT
MAV_4870|M.avium_104 KTVEALGLSGSPLPIPWSVLSLFAKGPTLSRADALLAHDGLPPDQAPGRV
MAB_2233c|M.abscessus_ATCC_199 KTLEALGLHKPALTIPSSILDLFASGPSLSRADALLARDSLTPTDRQAVP
MSMEG_0616|M.smegmatis_MC2_155 KVVAALGLSTSPLPVPWSVLSQFAAGPALSRGDALVAHDAVSTNPNSARM
TH_2593|M.thermoresistible__bu DTVAALGLTGAPLPMPWAVLAQFAAGPTLSRTDALLAHDSLPPNPAPARL
Mflv_0328|M.gilvum_PYR-GCK DTVAALGLTTTPLPIPSSVLSLFAAGPTLSRGDALLAHDTLAPNPSPARV
Mvan_0412|M.vanbaalenii_PYR-1 DTVAALGLTTAPLPIPSSVLSQFAVGPTLSRTDALLAHDSLAPNPAPARL
. : **** ..: :* ::* *. **:*** ***: :* : . .
MMAR_0542|M.marinum_M ASAE----GEPR---
MUL_1205|M.ulcerans_Agy99 ASAE----GGPR---
Mb0291|M.bovis_AF2122/97 VSAE----GGYR---
Rv0283|M.tuberculosis_H37Rv VSAE----GGYR---
MLBr_02536|M.leprae_Br4923 TTAY----GEHR---
MAV_4870|M.avium_104 AAAAPQAEGAPR---
MAB_2233c|M.abscessus_ATCC_199 VQTDTQLAQNAQESR
MSMEG_0616|M.smegmatis_MC2_155 EASR-----------
TH_2593|M.thermoresistible__bu ENHR---MENP----
Mflv_0328|M.gilvum_PYR-GCK EALPG--LENP----
Mvan_0412|M.vanbaalenii_PYR-1 ETTSA--LEKP----