For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSAIQVAAVDLGATSGRVMVADIDGDRLDMRTVARFPNDPVTLWNGTGDELRWNVPTLYGHVIDGLTRAG RDCDSLVGVGVDSWAVDYGLLREGRLLSLPMHYRDARTARGVELVHDVLDAPELYRRNGIQFLPFNTVYQ LAAERASGLLDLADTVLLVPDLLTYWLTGQRVAERTNASTTGLLALDGTWDRDLMTRLDLPHTLFPPIVE AGSRRGPLLPSVARQIGLTGKAGAEVVSVASHDTASAVAAIPMDPDHAAYISCGTWGLVGVELGRQVATD AAREANFTNEVGADQRIRFLHNVMGLWLLSETLRQYERDGHDVDLADLLAGAAQAPAPPVVFDTDDPRFL PPGDLPARISAWYVERGHTPPADPVQMVRAILESLAAAFASAVCTAAELSGVDVRTVHMVGGGSQNELLC QLTADRIGMPLLAGPVEATALGNVLLTARARHVISGDLASLRARVAQWFPPRRYLPRTSRTTAMV
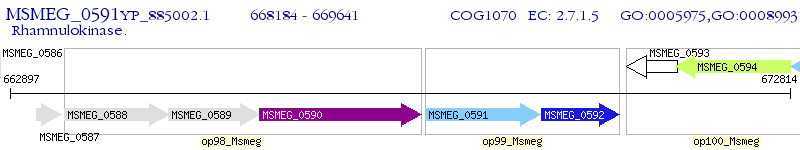
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0591 | rhaB | - | 100% (485) | rhamnulokinase |
| M. smegmatis MC2 155 | MSMEG_3113 | - | 4e-07 | 24.12% (452) | carbohydrate kinase, FGGY |
| M. smegmatis MC2 155 | MSMEG_3257 | xylB | 5e-05 | 22.91% (454) | xylulokinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_0570 | - | 2e-09 | 24.74% (481) | carbohydrate kinase |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_0046 | xylB | 1e-10 | 24.61% (451) | xylulokinase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0570|M.marinum_M --MTLVAGIDSSTQSCKVLVCDADTGEVARRGQAAHPDG-----------
TH_0046|M.thermoresistible__bu --MTLVAGIDSSTQSCKVLICDADTGAVVREGRAAHPDG-----------
MSMEG_0591|M.smegmatis_MC2_155 MSAIQVAAVDLGATSGRVMVADIDGDRLDMRTVARFPNDPVTLWNGTGDE
**.:* .: * :*::.* * . : . * .*:.
MMAR_0570|M.marinum_M -TEIDPAAWESAARAAIAAAGGFDDVEAVAVGAQQHGMVCLDMAGAVVRP
TH_0046|M.thermoresistible__bu -TEIDPAFWESAAHDAVRAAGGLDDVAAVAVGAQQHGMVCLDEAGKVVRD
MSMEG_0591|M.smegmatis_MC2_155 LRWNVPTLYGHVIDGLTRAGRDCDSLVGVGVDSWAVDYGLLREG-RLLSL
*: : . *. . *.: .*.*.: . * . ::
MMAR_0570|M.marinum_M ALLWNDVRSADAALALIDELGGPRAWAQAVGLVPLAAVTVAKLRWLADNE
TH_0046|M.thermoresistible__bu ALLWNDVRSAGAAAALVDELGGPQAWAEAVGVVPLAAITVAKLRWLADHE
MSMEG_0591|M.smegmatis_MC2_155 PMHYRDARTARGVELVHDVLDAPELYRRN-GIQFLPFNTVYQL--AAERA
.: :.*.*:* .. : * *..*. : . *: *. ** :* *:.
MMAR_0570|M.marinum_M PQHADRTAAVCLPHDWLTWRLRGDTDVATLTTDRSDASGTGYYDAGSDRY
TH_0046|M.thermoresistible__bu PQHADRTAAVCLPHDWLTWRLRGSTDIAELTTDRSDASGTGYYDASGDDY
MSMEG_0591|M.smegmatis_MC2_155 SGLLDLADTVLLVPDLLTYWLTGQRVAERTNASTTGLLALDGTWDRDLMT
. * : :* * * **: * *. .:. :. . . .
MMAR_0570|M.marinum_M RFDLLELAMH-------GRRPNLPRVLGPSARA-TRGSKTFGAGLGDNAA
TH_0046|M.thermoresistible__bu RLDLLELGLR-------GRRPHLPAVLGPSDAT-RGGDRVFGAGLGDNAA
MSMEG_0591|M.smegmatis_MC2_155 RLDLPHTLFPPIVEAGSRRGPLLPSVARQIGLTGKAGAEVVSVASHDTAS
*:** . : * * ** * : * ...... *.*:
MMAR_0570|M.marinum_M AALGLGADEG-DVMVSIGTSGVVCAVAADPVRDETGLVAGFADGTG--RY
TH_0046|M.thermoresistible__bu AALGLAAEPG-DVIVSVGTSGVVSAVATAPVRDGTGFVAGFADGTG--RH
MSMEG_0591|M.smegmatis_MC2_155 AVAAIPMDPDHAAYISCGTWGLVGVELGRQVATDAAREANFTNEVGADQR
*. .: : . . :* ** *:* . * :. *.*:: .* :
MMAR_0570|M.marinum_M LPLVCTLNGARVLDAAAAMLRVDHDELSRLALSAPPGSAGLVLVPYLEGE
TH_0046|M.thermoresistible__bu LPLVCTLNGARVLDAAAAMLQVDHAELSRLALSAPPGSEGLVLVPYLEGE
MSMEG_0591|M.smegmatis_MC2_155 IRFLHNVMGLWLLSETLRQYERDGHDVDLADLLAGAAQAPAPPVVFDTDD
: :: .: * :*. : . * ::. * * ... * : .:
MMAR_0570|M.marinum_M R----TPNRPKATGALHGLR--LANATPAHLARAAVEGLLCALADGLAQL
TH_0046|M.thermoresistible__bu R----TPNRPNASGALHGLR--VSNATPANLARAAVEGLLCALADGIAHL
MSMEG_0591|M.smegmatis_MC2_155 PRFLPPGDLPARISAWYVERGHTPPADPVQMVRAILESLAAAFASAVCTA
. : * .* : * . * *.::.** :*.* .*:*..:.
MMAR_0570|M.marinum_M TEHG-VVARRVLLIGGGARSQALREIAPLIFGVPVLVPDPAEYVALGAAR
TH_0046|M.thermoresistible__bu TGHG-IPLRRVLLIGGAARSEAVRAIAPQVFGVPVLVPDPAEYVALGAAR
MSMEG_0591|M.smegmatis_MC2_155 AELSGVDVRTVHMVGGGSQNELLCQLTADRIGMPLLAG-PVEATALGNVL
: . : * * ::**.::.: : ::. :*:*:*. *.* .*** .
MMAR_0570|M.marinum_M QAAWALRGSTRPPSWQPPATDEYTGEPSPAVRQRYGQVRELTEGVASVPG
TH_0046|M.thermoresistible__bu QAAWALRGTPEPPSWQPPRTDRYEADPVPAVRERYAEVRDLTEGVE----
MSMEG_0591|M.smegmatis_MC2_155 LTARARHVISGDLASLRARVAQWFPP------RRYLPRTSRTTAMV----
:* * : . : . . .: .** . * .:
MMAR_0570|M.marinum_M PGETPPAPDTSR
TH_0046|M.thermoresistible__bu ------------
MSMEG_0591|M.smegmatis_MC2_155 ------------