For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTSPQAHHAEIPAQSAGGATILEIGQQPDVWREIAGRSDAETSEFLRPIVERPDLRVILTGAGSSAFIGE IVAASLRRNLKRRVEAIATTDIVAVPLDHLERDTPTLLVSFGRSGNSPESVATTELADELVTDVWHLIFT CDPAGTLARTHEGRANSRVVHMPARTNDTGFAMTSSLTSMLLSCLLTLGGVRPESAEALAAAAEHVSGLS SDIRDLARAKKQRFVYLGSGPLTGLARESALKLLELTAGEVVTYFDSSLGFRHGPKSVLDSDTLAIVYVS TDPYTRRYDLDIIAELREQLGQEAVKAISAEPIPDALGPALVLPGLGGLDDAQVALPYLVFAQYLALFSS LEYGKTPDNPFPSGEVSRVVKGVTIHPMER
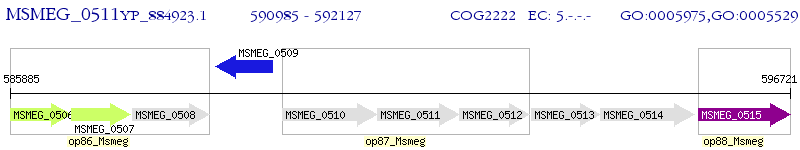
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0511 | - | - | 100% (380) | putative sugar isomerase, AgaS family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0363c | - | 6e-07 | 24.86% (358) | glutamine-fructose-6-phosphate transaminase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_0511|M.smegmatis_MC2_155 MTSPQAHHAEIPAQSAGGATILEIGQQPDVWREIAGRSDAETSEFLRPIV
MAB_0363c|M.abscessus_ATCC_199 -------------MTAGTHMADEIRQQPTVWGRLLSDGRSAIAEVAAKIV
:** ** *** ** .: . . : :*. **
MSMEG_0511|M.smegmatis_MC2_155 ERPDLRVILTGAGSSAFIGEIVAASLRRNLKRRVEAIATTDIVAVPLDHL
MAB_0363c|M.abscessus_ATCC_199 EYSPRFVLFVARGTSDHA-ALYGKYLTEIIWQLPAGLASPSTMTTYGARP
* . *::.. *:* . : . * . : : .:*:.. ::. :
MSMEG_0511|M.smegmatis_MC2_155 ERDTPTLLVSFGRSGNSPESVATTELADELVTDVWHLIFTCDPAGT-LAR
MAB_0363c|M.abscessus_ATCC_199 DLSG-VLVIGVSQSGGSPDLVQTLTVARQQG--------ACTVAVTNAAR
: . .*::...:**.**: * * :* : :* * * **
MSMEG_0511|M.smegmatis_MC2_155 THEGRANSRVVHMPARTNDTGFAMTSSLTSMLLSCLLTLGGVR--PESAE
MAB_0363c|M.abscessus_ATCC_199 SPLATAAEHHIDVLAGP-ELAVAATKSYTAQLLALYLLLTDVAGRDNTAA
: . * .: :.: * . : ..* *.* *: **: * * .* ::*
MSMEG_0511|M.smegmatis_MC2_155 ALAAAAEHVSGLSSDIRDLARAK--KQRFVYLGSGPLTGLARESALKLLE
MAB_0363c|M.abscessus_ATCC_199 ALLPERGEEILHSAVVQQLATRYRFASRLVTTGRGYSYPTAREAALKLME
** . . *: :::** .*:* * * ***:****:*
MSMEG_0511|M.smegmatis_MC2_155 LTAGEVVTYFDSSLGFRHGPKSVLDSDTLAIVYVSTDPYTRRYDLDIIAE
MAB_0363c|M.abscessus_ATCC_199 TSYLSAQAFSGADL--LHGPLAMVDHQVPVIAIVP-DGVGGRAMNDVLHR
: .. :: .:.* *** :::* :. .*. *. * * *:: .
MSMEG_0511|M.smegmatis_MC2_155 LREQLGQEAVKAISAEPIPDALGPALVLPGLGGLDDAQVALPYLVFAQYL
MAB_0363c|M.abscessus_ATCC_199 LAGRGADVCCIGSADAVAATGLG-VTLEPTLEELSPMLAIIPLQLLAMQL
* : .: . . : . .** . : * * *. . :* ::* *
MSMEG_0511|M.smegmatis_MC2_155 ALFSSLEYGKTPDNPFPSGEVSRVVKGVTIHPMER
MAB_0363c|M.abscessus_ATCC_199 AIGR----GENPDVPRGLSKVTETL----------
*: *:.** * .:*:..: