For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TAGTHMADEIRQQPTVWGRLLSDGRSAIAEVAAKIVEYSPRFVLFVARGTSDHAALYGKYLTEIIWQLPA GLASPSTMTTYGARPDLSGVLVIGVSQSGGSPDLVQTLTVARQQGACTVAVTNAARSPLATAAEHHIDVL AGPELAVAATKSYTAQLLALYLLLTDVAGRDNTAAALLPERGEEILHSAVVQQLATRYRFASRLVTTGRG YSYPTAREAALKLMETSYLSAQAFSGADLLHGPLAMVDHQVPVIAIVPDGVGGRAMNDVLHRLAGRGADV CCIGSADAVAATGLGVTLEPTLEELSPMLAIIPLQLLAMQLAIGRGENPDVPRGLSKVTETL*
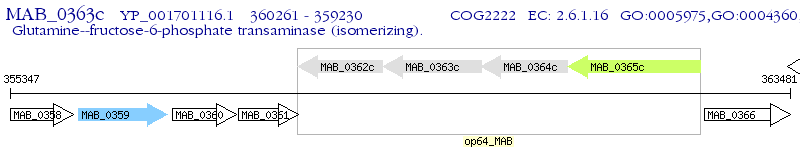
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_0363c | - | - | 100% (343) | glutamine-fructose-6-phosphate transaminase |
| M. abscessus ATCC 19977 | MAB_3743c | - | 8e-33 | 30.91% (317) | glucosamine--fructose-6-phosphate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3466c | glmS | 1e-32 | 29.12% (364) | glucosamine--fructose-6-phosphate aminotransferase |
| M. gilvum PYR-GCK | Mflv_4939 | - | 8e-31 | 29.34% (317) | D-fructose-6-phosphate amidotransferase |
| M. tuberculosis H37Rv | Rv3436c | glmS | 2e-32 | 29.12% (364) | glucosamine--fructose-6-phosphate aminotransferase |
| M. leprae Br4923 | MLBr_00371 | glmS | 5e-32 | 30.89% (314) | glucosamine--fructose-6-phosphate aminotransferase |
| M. marinum M | MMAR_1114 | glmS | 1e-32 | 30.14% (365) | glucosamine-fructose-6-phosphate aminotransferase, GlmS |
| M. avium 104 | MAV_4377 | glmS | 3e-32 | 30.75% (361) | D-fructose-6-phosphate amidotransferase |
| M. smegmatis MC2 155 | MSMEG_1568 | glmS | 2e-32 | 31.23% (317) | D-fructose-6-phosphate amidotransferase |
| M. thermoresistible (build 8) | TH_1701 | glmS | 2e-31 | 29.89% (358) | PROBABLE GLUCOSAMINE--FRUCTOSE-6-PHOSPHATE AMINOTRANSFERASE |
| M. ulcerans Agy99 | MUL_0872 | glmS | 9e-33 | 30.14% (365) | glucosamine--fructose-6-phosphate aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_1480 | - | 3e-31 | 29.65% (317) | D-fructose-6-phosphate amidotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4939|M.gilvum_PYR-GCK MAMCGIVGYVGQRPACDIVVDALRRMEYRGYDSSGVALVDGH---GGLTV
Mvan_1480|M.vanbaalenii_PYR-1 --MCGIVGYVGQRPACDIVVDALRRMEYRGYDSAGVALLDGR---GGLTV
MSMEG_1568|M.smegmatis_MC2_155 --MCGIVGYVGHRPARDIVVDALRRMEYRGYDSAGIALIDGN---GGLTV
TH_1701|M.thermoresistible__bu --MCGIVGYVGARNALDIVVDALRRMEYRGYDSAGVALLDGA---GGLTV
Mb3466c|M.bovis_AF2122/97 --MCGIVGYVGRRPAYVVVMDALRRMEYRGYDSSGIALVDG----GTLTV
Rv3436c|M.tuberculosis_H37Rv --MCGIVGYVGRRPAYVVVMDALRRMEYRGYDSSGIALVDG----GTLTV
MMAR_1114|M.marinum_M --MCGIVGYVGQRPACEVVLDALRRMEYRGYDSSGIALVDGS---GKLTV
MUL_0872|M.ulcerans_Agy99 --MCGIVGYVGQRPACEVVLDALRRMEYRGYDSSGIALVDGS---GKLTV
MLBr_00371|M.leprae_Br4923 --MCGLVGYVGQRPACGVVMDALRRMEYRGYDSSGIALINGSAKSGNLTV
MAV_4377|M.avium_104 --MCGIVGYVGQQPACAVVMAALRRMEYRGYDSSGVALVNGD---GTLTV
MAB_0363c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4939|M.gilvum_PYR-GCK RRKAGRLANLEAALAEAG---AESPAGATGLGHTRWATHGRPTDRNAHPH
Mvan_1480|M.vanbaalenii_PYR-1 RRKAGRLANLEAALAEGG---TDNLVGATGLGHTRWATHGRPTDRNAHPH
MSMEG_1568|M.smegmatis_MC2_155 RRRAGRLANLEATLAETDSNDGDGLGGSTGLGHTRWATHGRPTDRNAHPH
TH_1701|M.thermoresistible__bu RRRAGRLANLEAELAATD---PGLLAGTAGLGHTRWATHGRPTDRNAHPH
Mb3466c|M.bovis_AF2122/97 RRRAGRLANLEEAVAEMP---STALSGTTGLGHTRWATHGRPTDRNAHPH
Rv3436c|M.tuberculosis_H37Rv RRRAGRLANLEEAVAEMP---STALSGTTGLGHTRWATHGRPTDRNAHPH
MMAR_1114|M.marinum_M RRRAGRLANLETAVAEMS---AESLTGTTGLGHTRWATHGRPTDRNAHPH
MUL_0872|M.ulcerans_Agy99 RRRAGRLANLETAVAEMP---AESLTGTTGLGHTRWATHGRPTDRNAHPH
MLBr_00371|M.leprae_Br4923 RRRAGRLSNLESVLAEMV---PASLAGNVGLGHIRWATHGRPTDRNAHPH
MAV_4377|M.avium_104 SRRAGRLANLEEAVAQLP---PSALTGTTGLGHTRWATHGRPTDRNAHPH
MAB_0363c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4939|M.gilvum_PYR-GCK RDSSGKLAVVHNGIIENFAALRAELEAVGVEFASDTDTEVAVHLVAREYT
Mvan_1480|M.vanbaalenii_PYR-1 RDASGKVAVVHNGIIENFAVLRAELEAAGVEFASDTDTEVTVHLVAREYV
MSMEG_1568|M.smegmatis_MC2_155 RDAAGKIAVVHNGIIENFAPLRAELEAAGVEFASDTDTEVAVHLVARQYT
TH_1701|M.thermoresistible__bu RDASGKFAVAHNGIIENNAVLRAELEAAGVEFASDTDTEVAVHLLARAYQ
Mb3466c|M.bovis_AF2122/97 RDAAGKIAVVHNGIIENFAVLRRELETAGVEFASDTDTEVAAHLVARAYR
Rv3436c|M.tuberculosis_H37Rv RDAAGKIAVVHNGIIENFAVLRRELETAGVEFASDTDTEVAAHLVARAYR
MMAR_1114|M.marinum_M RDAAGKIAVVHNGIIENFAVLRQELEAAGVEFASDTDTEVAVHLVAQAFH
MUL_0872|M.ulcerans_Agy99 RDAAGKIAVVHNGIIENFAVLRQELEAAGVEFASDTDTEVAVHLVAQAFH
MLBr_00371|M.leprae_Br4923 RDATGKIAVVHNGIIENFPSLRHELEIAGVEFVSDTDTEVAVHLVAQAYC
MAV_4377|M.avium_104 RDAAGKIAVVHNGIIENYAGLRHELEADGVEFASDTDTEVAVHLVAQAYR
MAB_0363c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4939|M.gilvum_PYR-GCK HGDTAGDFPASVFAVLPRLEGHFTLVFAHADDPGTIVAARRSTPLVLGVG
Mvan_1480|M.vanbaalenii_PYR-1 HGETAGDFPASVLAVLRRLEGHFTLVFAHADDPGTIVAARRSTPLVLGVG
MSMEG_1568|M.smegmatis_MC2_155 QGDTAGDFPASVLAVLQRLEGHFTLVFASADDPGTIVAARRSTPLVLGIG
TH_1701|M.thermoresistible__bu HGETAGDFVASVLSVLRRLDGHFTLVFANADEPGTIIAARRSTPLVVGVG
Mb3466c|M.bovis_AF2122/97 HGETADDFVGSVLAVLRRLEGHFTLVFANADDPGTLVAARRSTPLVLGIG
Rv3436c|M.tuberculosis_H37Rv HGETADDFVGSVLAVLRRLEGHFTLVFANADDPGTLVAARRSTPLVLGIG
MMAR_1114|M.marinum_M HGETAGDFPASVLSVLRRLEGHFTLVFANADDPGTIVAARRSTPLVLGIG
MUL_0872|M.ulcerans_Agy99 HGETAGDFPASVLSVLRRLEGHFTLVFANADDPGTIVAARRSTPLVLGIG
MLBr_00371|M.leprae_Br4923 AGETAGDFVGSVLAVLRRLQGHFTLVFANADEPGTIVAARRSTPLVLGIG
MAV_4377|M.avium_104 HGLTAGDFAASVLAVLRRLDGHFTLVFANADEPGTIVAARRSTPLVIGIG
MAB_0363c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4939|M.gilvum_PYR-GCK DGEMFVGSDVAAFIEHTREAVELGQDQAVVVTADGYRVTDFHGNDDAAS-
Mvan_1480|M.vanbaalenii_PYR-1 DGEMFVGSDVAAFIEHTRDAVELGQDQAVVVTADGYRVTDFHGNPDDAR-
MSMEG_1568|M.smegmatis_MC2_155 DGEMFVGSDVAAFIEHTRDAVELGQDQAVVLTADGYRITDFAGNDHLEAG
TH_1701|M.thermoresistible__bu DGEMFVGSDVAAFIEHTRDAVELGQDQAVVITADSYRVLDFLGADVTAQ-
Mb3466c|M.bovis_AF2122/97 DNEMFVGSDVAAFIEHTREAVELGQDQAVVITADGYRISDFDGNDGLQAG
Rv3436c|M.tuberculosis_H37Rv DNEMFVGSDVAAFIEHTREAVELGQDQAVVITADGYRISDFDGNDGLQAG
MMAR_1114|M.marinum_M DGEMFVGSDVAAFITHTRHAVELGQDQAVVITADGYRISDFDGNDDLLAG
MUL_0872|M.ulcerans_Agy99 DGEMFVGSDVAAFITHTRHAVDLGQDQAVVITADGYRISDFDGNDDLLAG
MLBr_00371|M.leprae_Br4923 DGEMFVGSDVAAFIEHTRQAVELGQDQAVVITADGYRISDFDGNDDAVN-
MAV_4377|M.avium_104 DGEMFVGSDVAAFIPHTRNAIELGQDQAVVLTADGYRITDFDGNDDLGPG
MAB_0363c|M.abscessus_ATCC_199 ------------------------------MTAG----------------
:**.
Mflv_4939|M.gilvum_PYR-GCK --ARPFHIDWDLSAAEKGGYEYFMLKEIAEQPAAVSDTLLGHFVDNRIVL
Mvan_1480|M.vanbaalenii_PYR-1 --TRRFHIDWDTSAAEKGGYEYFMLKEIAEQPAAVSDTLLGHFADNRIVL
MSMEG_1568|M.smegmatis_MC2_155 RDFREFHIDWDLNAAEKGGYDYFMLKEIAEQPSAVADTLLGHFDKNRIVL
TH_1701|M.thermoresistible__bu --ARHFHIDWDLSAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVDGRIVL
Mb3466c|M.bovis_AF2122/97 RDFRPFHIDWDLAAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVGGRIVL
Rv3436c|M.tuberculosis_H37Rv RDFRPFHIDWDLAAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVGGRIVL
MMAR_1114|M.marinum_M TDYREFHIDWDLAAAEKGGYEYFMLKEIAEQPTAVAETLLGHFVGNRIVL
MUL_0872|M.ulcerans_Agy99 TDYREFHIDWDLAAAEKGGYEYFMLKEIAEQPTAVAETLLGHFVGNRIVL
MLBr_00371|M.leprae_Br4923 --ARTFHIDWDLAAAEKGGYEYFMLKEIAEQPDAVVDTLLGHFTGGRIVL
MAV_4377|M.avium_104 -AYREFHIDWDLAAAEKGGYEYFMLKEIAEQPAAVADTLLGHFVDGRIVL
MAB_0363c|M.abscessus_ATCC_199 ---------------------THMADEIRQQP-----TVWGRLLSDGRSA
.* .** :** *: *:: .
Mflv_4939|M.gilvum_PYR-GCK DEQRLSDQELRDIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
Mvan_1480|M.vanbaalenii_PYR-1 DEQRLSDQELRDIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MSMEG_1568|M.smegmatis_MC2_155 DEQRLSDQELREIDKVFIVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
TH_1701|M.thermoresistible__bu DEQRLSDQELREVDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
Mb3466c|M.bovis_AF2122/97 DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
Rv3436c|M.tuberculosis_H37Rv DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MMAR_1114|M.marinum_M DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MUL_0872|M.ulcerans_Agy99 DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MLBr_00371|M.leprae_Br4923 DEQRLSDQELREIDKVFVVACGTAYHSGLLAKYTIEHWTRLPVEVELASE
MAV_4377|M.avium_104 DEQRLSDQELREVDKVFVVACGTAYHSGLLAKYAIEHWTRLPVEVELASE
MAB_0363c|M.abscessus_ATCC_199 IAEVAAKIVEYSPRFVLFVARGTSDHAALYGKYLTEIIWQLPAGLASPST
: :. . *:.** **: *:.* .** * :**. : .*
Mflv_4939|M.gilvum_PYR-GCK FRYRDPVLDSH-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
Mvan_1480|M.vanbaalenii_PYR-1 FRYRDPVLDSH-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
MSMEG_1568|M.smegmatis_MC2_155 FRYRDPVLDRS-TLVIAISQSGETADTLEAVRHAKTQKAKVLAICNTNGS
TH_1701|M.thermoresistible__bu FRYRDPVLDRS-TLVVAISQSGETADTLEAVRHAKAQKAKVLAICNTNGS
Mb3466c|M.bovis_AF2122/97 FRYRDPVLDRS-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
Rv3436c|M.tuberculosis_H37Rv FRYRDPVLDRS-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
MMAR_1114|M.marinum_M FRYRDPVLDRS-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
MUL_0872|M.ulcerans_Agy99 FRYRDPVLDRS-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
MLBr_00371|M.leprae_Br4923 FRYRDPVLDRS-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
MAV_4377|M.avium_104 FRYRDPVLDRS-TLVVAISQSGETADTLEAVRHAKEQKAKVLAICNTNGS
MAB_0363c|M.abscessus_ATCC_199 MTTYGARPDLSGVLVIGVSQSGGSPDLVQTLTVARQQGACTVAVTNAARS
: .. * .**:.:**** :.* :::: *: * * .:*: *: *
Mflv_4939|M.gilvum_PYR-GCK QIPRECDAVLYTRAGPEIGVASTKTFLAQVAANYLVGLALAQARGTKYPD
Mvan_1480|M.vanbaalenii_PYR-1 QIPRECDAVLYTRAGPEIGVASTKTFLAQIAANYLVGLALAQARGTKYPD
MSMEG_1568|M.smegmatis_MC2_155 QIPREADAVLYTRAGPEIGVAATKTFLAQIAANYLVGLALAQARGTKYPD
TH_1701|M.thermoresistible__bu QIPRESDAVLYTRAGPEIGVAATKTFLAQIVANYLVGLALAQARGTKYPD
Mb3466c|M.bovis_AF2122/97 QIPRECDAVLYTRAGPEIGVASTKTFLAQIAANYLLGLALAQARGTKYPD
Rv3436c|M.tuberculosis_H37Rv QIPRECDAVLYTRAGPEIGVASTKTFLAQIAANYLLGLALAQARGTKYPD
MMAR_1114|M.marinum_M QIPRECDAVLYTRAGPEIGVASTKTFLAQVTANYLLGLALAQARGTKYPD
MUL_0872|M.ulcerans_Agy99 QIPRECDAVLYTRAGPEIGVASTKTFLAQVTANYLLGLALAQARGTKYPD
MLBr_00371|M.leprae_Br4923 QIPRECDAVLYTRAGPEIGVASTKTFLAQVAANYLLGLALAQARGTKYPD
MAV_4377|M.avium_104 QIPRECDAVLYTRAGPEIGVASTKTFLAQITANYLVGLALAQARGTKYPD
MAB_0363c|M.abscessus_ATCC_199 PLATAAEHHIDVLAGPELAVAATKSYTAQLLALYLL-----------LTD
:. .: : . ****:.**:**:: **: * **: .*
Mflv_4939|M.gilvum_PYR-GCK EVAREYHDLEAMPDLISRVLASSEPVVALAKQFATSQTVLFLGRHVGYPV
Mvan_1480|M.vanbaalenii_PYR-1 EVAREYHDLEAMPDLIARVLDTSEPVLQLAKQFAASQTVLFLGRHVGYPV
MSMEG_1568|M.smegmatis_MC2_155 EVAREYRELEAMPDLIKRVLAGMDSVAALAERFAPSSTVLFLGRHVGYPV
TH_1701|M.thermoresistible__bu EVEREYRELEAMPALVQRVVSTVEPVSALARQFAGAPAVLFLGRHVGYPV
Mb3466c|M.bovis_AF2122/97 EVEREYHELEAMPDLVARVIAATGPVAELAHRFAQSSTVLFLGRHVGYPV
Rv3436c|M.tuberculosis_H37Rv EVEREYHELEAMPDLVARVIAATGPVAELAHRFAQSSTVLFLGRHVGYPV
MMAR_1114|M.marinum_M EVEREYRELEAMPDLVARVISAIKPVADLAYRFAQSSTVLFLGRHVGYPV
MUL_0872|M.ulcerans_Agy99 EVEREYRELEAMPDLVARVISAIKPVAHLAYRFAQSSTVLFLGRHVGYPV
MLBr_00371|M.leprae_Br4923 EVQREYRELEAMPDLVARVIAGMGPVADLAYRFAQSTTVLFLGRHVGYPV
MAV_4377|M.avium_104 EVEREYHELEAMPDLVARVLATIKPVAALAQRFAQSPTVLFLGRHVGYPV
MAB_0363c|M.abscessus_ATCC_199 VAGRDNTAAALLPERGEEILHS-AVVQQLATRYRFASRLVTTGRGYSYPT
. *: :* .:: * ** :: : :: ** .**.
Mflv_4939|M.gilvum_PYR-GCK ALEGALKLKELAYMHAEGFAAGELKHGPIALIEDDLPVIVVMPSPKNSAT
Mvan_1480|M.vanbaalenii_PYR-1 ALEGALKLKELAYMHAEGFAAGELKHGPIALIEDDLPVIVVMPSPKNSAT
MSMEG_1568|M.smegmatis_MC2_155 ALEGALKLKELAYMHAEGFAAGELKHGPIALIDENLPVIVVMPSPKNAAM
TH_1701|M.thermoresistible__bu ALEGALKLKELAYMHAEGFAAGELKHGPIALIEEGLPVIVVMPSPKHALT
Mb3466c|M.bovis_AF2122/97 ALEGALKLKELAYMHAEGFAAGELKHGPIALIEDGLPVIVVMPSPKGSAT
Rv3436c|M.tuberculosis_H37Rv ALEGALKLKELAYMHAEGFAAGELKHGPIALIEDGLPVIVVMPSPKGSAT
MMAR_1114|M.marinum_M ALEGALKLKELAYMHAEGFAAGELKHGPIALVEDELPVIVIMPSPKGSAT
MUL_0872|M.ulcerans_Agy99 ALEGALKLKELAYMHAEGFAAGELKHGPIALVEDELPVIVIMPSPKGSAT
MLBr_00371|M.leprae_Br4923 ALEGALKLKELAYMHAEGFAAGELKHGPIALIEENLPVIVVMPSPKGSAM
MAV_4377|M.avium_104 ALEGALKLKELAYMHAEGFAAGELKHGPIALIEDDLPVIVIMPSPKGSAV
MAB_0363c|M.abscessus_ATCC_199 AREAALKLMETSYLSAQAFSGADLLHGPLAMVDHQVPVIAIVPDGVGGRA
* *.**** * :*: *:.*:..:* ***:*:::. :***.::*. .
Mflv_4939|M.gilvum_PYR-GCK LHSKLLSNIREIQARGAVTIVIAEEGDETVRPYADHLIEIPAVSTLFQPL
Mvan_1480|M.vanbaalenii_PYR-1 LHSKLLSNIREIQARGAVTIVIAEEGDDTVRPYADHLIEIPAVSTLFQPL
MSMEG_1568|M.smegmatis_MC2_155 LHAKLLSNIREIQARGAVTVVIAEEDDDTVRPYADHLIEIPSVSTLFQPL
TH_1701|M.thermoresistible__bu LHGKLLSNIREIQARGAVTIVIAEEGDDTVRPYADHLIEIPVVSTLFQPL
Mb3466c|M.bovis_AF2122/97 LHAKLLSNIREIQTRGAVTIVIAEEGDETVRPYADHLIEIPAVSTLLQPL
Rv3436c|M.tuberculosis_H37Rv LHAKLLSNIREIQTRGAVTIVIAEEGDETVRPYADHLIEIPAVSTLLQPL
MMAR_1114|M.marinum_M LHAKLLSNIREIQTRGAVTIVIAEEGDDTVRPFADHLIEIPAVSTLLQPL
MUL_0872|M.ulcerans_Agy99 LHAKLLSNIREIQTRGAVTIVIAEEGDDTVRPFADHLIEIPAVSTLLQPL
MLBr_00371|M.leprae_Br4923 LHAKLLSNIREIQTRGAVTIVIAEEGDDTVRLYADHLIELPAVSTLLQPL
MAV_4377|M.avium_104 LHAKLLSNIREIQARGAITIVIAEEGDDTVRPYADHLIEIPSVSTLLQPL
MAB_0363c|M.abscessus_ATCC_199 MNDVLHR----LAGRGADVCCIGSADAVAATGLG---VTLEPTLEELSPM
:: * : *** . *.. . :. . : : . :.*:
Mflv_4939|M.gilvum_PYR-GCK LSTIPMQLFSAGVAQARGYDVDKPRNLAKSVTVE
Mvan_1480|M.vanbaalenii_PYR-1 LSTIPMQLFSAGVAQARGYDVDKPRNLAKSVTVE
MSMEG_1568|M.smegmatis_MC2_155 LSTIPLQVFAAGVARARGYDVDKPRNLAKSVTVE
TH_1701|M.thermoresistible__bu LSTVPLQVFAASVAQARGYDVDKPRNLAKSVTVE
Mb3466c|M.bovis_AF2122/97 LSTIPLQVFAASVARARGYDVDKPRNLAKSVTVE
Rv3436c|M.tuberculosis_H37Rv LSTIPLQVFAASVARARGYDVDKPRNLAKSVTVE
MMAR_1114|M.marinum_M LSTIPLQVFAAAVAQARGYDVDKPRNLAKSVTVE
MUL_0872|M.ulcerans_Agy99 LSTIPLQVFAAAVAQARGYDVDKPRNLAKSVTVE
MLBr_00371|M.leprae_Br4923 LSTIPLQVFAASVAQARGYDVDKPRNLAKSVTVE
MAV_4377|M.avium_104 LSTIPLQVFAASVAQARGYDVDKPRNLAKSVTVE
MAB_0363c|M.abscessus_ATCC_199 LAIIPLQLLAMQLAIGRGENPDVPRGLSKVTETL
*: :*:*::: :* .** : * **.*:* . .