For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TATLDVPEQNPDLVLDQSADDYWNHYQLTFALYSVSDRAIPSAYDGLKPGQRRLLYQMHDSKLLPGNKPQ KSSKVCSAVTGNLHPHGGASMYGAAALMAADFQRVKVIDGQGAFPRIQGDIPAADRYTEMRLSAPGAALT AELNDHAVPMVPTFDGEWVEPTVLPAQWPVLLCNGAVGIAEGWATKVPAHNPREVMAACRALLKTPNMTD DRLCKLIPGPDWGSGASVVGTAGLREYITTGRGHFTVRGTISVEGKNCIITELPPGVASNTVQDRIRALV ESGEMSGVADMSDLTDRRNGLRIVVTAKRGHNAEQIRDQLLALTPLESTFAASLVALDENRVPRWWSVRD LIMAFLQLRDSVVLHRSEYRLEKVTARRHLVAGLMKIHLDIDAAVAVIRGSETVDEARKGLQERFKIDAE QADYVLSLQLRRLTKLDVIELQAEAEKLDAEFAELNDLVTNPESRRKVIDKELVETAKLFKGPEYDRRTV LDFDATPVSRGDEDGSRERKVNTAWRLDDRGVFSDSHGELLTSGLGWAVWSDGRIKFTTGNGLPFKIRDI PVAPDITGLVRSGVLPEGYHLALVTRRGKILRVDPASVNPQGVAGNGVAGVKLAADGDEVIAALPVSCAN GEAILSIAEKSWKVTEVADIPVKGRGGAGVAFHPFVKGEDALLAASISKTGYVRGKRAVRPENRAKASIK GSGADVTPAPAE*
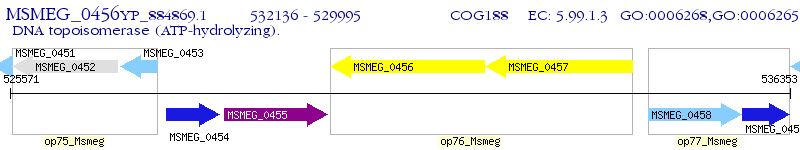
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0456 | - | - | 100% (713) | DNA gyrase subunit A |
| M. smegmatis MC2 155 | MSMEG_0006 | gyrA | 1e-78 | 36.46% (458) | DNA gyrase subunit A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0006 | gyrA | 2e-78 | 30.30% (726) | DNA gyrase subunit A |
| M. gilvum PYR-GCK | Mflv_0497 | - | 0.0 | 88.11% (715) | DNA gyrase/topoisomerase IV, subunit A |
| M. tuberculosis H37Rv | Rv0006 | gyrA | 2e-79 | 30.36% (728) | DNA gyrase subunit A |
| M. leprae Br4923 | MLBr_00006 | gyrA | 9e-54 | 27.78% (594) | putative DNA gyrase subunit A |
| M. abscessus ATCC 19977 | MAB_0019 | - | 3e-82 | 31.23% (698) | DNA gyrase subunit A |
| M. marinum M | MMAR_0006 | gyrA | 6e-79 | 30.29% (700) | DNA gyrase (subunit A) GyrA |
| M. avium 104 | MAV_0006 | gyrA | 2e-79 | 30.80% (698) | DNA gyrase subunit A |
| M. thermoresistible (build 8) | TH_0756 | gyrA | 9e-81 | 30.95% (727) | DNA GYRASE (SUBUNIT A) GYRA (DNA TOPOISOMERASE |
| M. ulcerans Agy99 | MUL_0006 | gyrA | 4e-79 | 30.29% (700) | DNA gyrase subunit A |
| M. vanbaalenii PYR-1 | Mvan_5898 | - | 0.0 | 90.60% (713) | DNA gyrase/topoisomerase IV, subunit A |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0006|M.marinum_M MTDTTLPPGG-EASDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MUL_0006|M.ulcerans_Agy99 MTDTMLPPGG-EASDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
Mb0006|M.bovis_AF2122/97 MTDTTLPPD--DSLDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
Rv0006|M.tuberculosis_H37Rv MTDTTLPPD--DSLDRIEPVDIEQEMQRSYIDYAMSVIVGRALPEVRDGL
MAV_0006|M.avium_104 MTDTTLPPGG-DAADRVEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MLBr_00006|M.leprae_Br4923 MTDITLPPGD-GSIQRVEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MAB_0019|M.abscessus_ATCC_1997 MTDTTLPPGGDDAVDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
TH_0756|M.thermoresistible__bu MTDTTLPPGG-EAGDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MSMEG_0456|M.smegmatis_MC2_155 -MTATLDVPE-QNPDLVLDQSADDYWNHYQLTFALYSVSDRAIPSAYDGL
Mvan_5898|M.vanbaalenii_PYR-1 -MTATLDVPE-QNPDLVLDQSADDYWNQYQLTFALYSVSDRAIPSAFDGL
Mflv_0497|M.gilvum_PYR-GCK -MTATIE----QNPELVLDQSADEYWNHYQLTFALYSVSDRAIPSAFDGL
: : : . :: :: : :*: : .**:*.. ***
MMAR_0006|M.marinum_M KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MUL_0006|M.ulcerans_Agy99 KPVHRRVLYAMYDSGFRPDHSHAKSARSVAETMGNYHPHGDASIYDTLVR
Mb0006|M.bovis_AF2122/97 KPVHRRVLYAMFDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
Rv0006|M.tuberculosis_H37Rv KPVHRRVLYAMFDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDSLVR
MAV_0006|M.avium_104 KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MLBr_00006|M.leprae_Br4923 KPVHRRVLYAMLDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MAB_0019|M.abscessus_ATCC_1997 KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
TH_0756|M.thermoresistible__bu KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MSMEG_0456|M.smegmatis_MC2_155 KPGQRRLLYQMHDSKLLPGNKPQKSSKVCSAVTGNLHPHGGASMYGAAAL
Mvan_5898|M.vanbaalenii_PYR-1 KPGQRRLLYQMHDSKLLPGNKPQKSSKICSAVTGNLHPHGGASMYGAAAL
Mflv_0497|M.gilvum_PYR-GCK KPGQRRLLYQMHESRLLPGNKPQKSSKICSAVTGNLHPHGGASMYGAAAL
** :**:** * :* : *... **:: : . ** ****.**:*.: .
MMAR_0006|M.marinum_M MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MUL_0006|M.ulcerans_Agy99 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
Mb0006|M.bovis_AF2122/97 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
Rv0006|M.tuberculosis_H37Rv MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MAV_0006|M.avium_104 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MLBr_00006|M.leprae_Br4923 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRYCVSGNSLVRLLFGKSIRIG
MAB_0019|M.abscessus_ATCC_1997 MAQPWSLRYPLVDGQGNFGSPGNDPAAAMRY-------------------
TH_0756|M.thermoresistible__bu MAQPWSMRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MSMEG_0456|M.smegmatis_MC2_155 MAADFQR-VKVIDGQGAFPRIQGDIPAADRY-------------------
Mvan_5898|M.vanbaalenii_PYR-1 MAAEFQR-VKVIDGQGAFPRIQGDIPAADRY-------------------
Mflv_0497|M.gilvum_PYR-GCK MAADFQR-VKVIDGQGAFPRIQGDIPAADRY-------------------
** :. ::**** * .* .** **
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 DIVTGAQFNSDNPIDLKVLDRHGNPVVADYLFHSGEHQTYTVRTTEGYEI
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 TGTSNHPLLCLVNVGGIPTLLWKLIGEIRSGDYVVLQRIPPVEFGPADWY
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 STMEALLFGAFISGGFVFQDHAGFNSLDRDYFTMVVNAYDTVVGGLRCIS
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 SRITVSGSTLLELDVYNLIEFKKTRLSGLCGQRSADKLVPDWLWHSPSTV
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 KRAFLQALFEGEGFSSILSRNIIEISYSTPSERLAADVQQMLLEFGVVSE
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 RYCHTVNEYKVVIANRAQVEMFFTQVGFGVTKQAKLIRDVVSMSPCVGMD
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 INCVPGLATFIRKHCDNRWVEEDSFNQHNVDCVQHWHHHSAEIVGHIADP
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0006|M.avium_104 --------------------------------------------------
MLBr_00006|M.leprae_Br4923 DIRAIVTDLTDGRFYYARVASVTDTGIQPVFSLHVDTEDHSFLTNGFISH
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0456|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_5898|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_0497|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_0006|M.marinum_M -TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANGS
MUL_0006|M.ulcerans_Agy99 -TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANGS
Mb0006|M.bovis_AF2122/97 -TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANGS
Rv0006|M.tuberculosis_H37Rv -TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANGS
MAV_0006|M.avium_104 -TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANGS
MLBr_00006|M.leprae_Br4923 NTEARLTPLAMEMLREIDEETVDFISNYDGRVQEPMVLPSRFPNLLANGS
MAB_0019|M.abscessus_ATCC_1997 -TEARLTPLAMEMLREIDEETVDFIPNYDGRVMEPTVLPSRFPNLLANGS
TH_0756|M.thermoresistible__bu -TEARLTPLAMEMLREIDEETVDFVPNYDGRVQEPTVLPSRFPNLLANGS
MSMEG_0456|M.smegmatis_MC2_155 -TEMRLSAPGAALTAELNDHAVPMVPTFDGEWVEPTVLPAQWPVLLCNGA
Mvan_5898|M.vanbaalenii_PYR-1 -TEMRLSAPGAALTAELDNHAVPMVPTFDGEWTEPTWLPARWPVLLCNGA
Mflv_0497|M.gilvum_PYR-GCK -TEMRLSAPGAALTAELDSHAVPMVATFDGEWTEPTVLPAQWPVLLCNGA
** **:. . : *::..:* ::..:**. ** **:::* **.**:
MMAR_0006|M.marinum_M GGIAVGMATNIPPHNLRELAEAVFWCLENHDADEEATLAAVTERVKGPDF
MUL_0006|M.ulcerans_Agy99 GGIAVGMATNIPPHNLRELAEAVFWCLENHDADEEATLAAVTDRVKGPDF
Mb0006|M.bovis_AF2122/97 GGIAVGMATNIPPHNLRELADAVFWALENHDADEEETLAAVMGRVKGPDF
Rv0006|M.tuberculosis_H37Rv GGIAVGMATNIPPHNLRELADAVFWALENHDADEEETLAAVMGRVKGPDF
MAV_0006|M.avium_104 GGIAVGMATNIPPHNLGELAEAVFWALDNYEADEEATLAAVMERVKGPDF
MLBr_00006|M.leprae_Br4923 GGIAVGMATNIPPHNLYELADAVFWCLENHDADEETMLVAVMERVKGPDF
MAB_0019|M.abscessus_ATCC_1997 GGIAVGMATNMPPHNLRELAEAVYWALDNHEADEETTLKAVCEKITGPDF
TH_0756|M.thermoresistible__bu GGIAVGMATNIPPHNLRELAEAVYWCLENYEADEETTLAAVMERVKGPDF
MSMEG_0456|M.smegmatis_MC2_155 VGIAEGWATKVPAHNPREVMAACRALLKTPNMTDDR----LCKLIPGPDW
Mvan_5898|M.vanbaalenii_PYR-1 VGIAEGWATKVPAHNPREVMAACRALLKTPNMTDDR----LVKLIPGPDW
Mflv_0497|M.gilvum_PYR-GCK VGIAEGWATKVPAHNPREVMAACRALLKTPNMTDDR----LVKLIPGPDW
*** * **::*.** *: * *.. : :: : : ***:
MMAR_0006|M.marinum_M PTSGLIVGTQGISDAYKTGRGSIRMRGVVEIEEDSRGRTSLVITELPYQV
MUL_0006|M.ulcerans_Agy99 PTSGLIVGTQGISDAYKTGRGSIRMRGVVEIEEDSRGRTSLVITELPYQV
Mb0006|M.bovis_AF2122/97 PTAGLIVGSQGTADAYKTGRGSIRMRGVVEVEEDSRGRTSLVITELPYQV
Rv0006|M.tuberculosis_H37Rv PTAGLIVGSQGTADAYKTGRGSIRMRGVVEVEEDSRGRTSLVITELPYQV
MAV_0006|M.avium_104 PTSGLIVGTQGIADAYKTGRGSIRMRGVVEVEEDSRGRTSLVITELPYQV
MLBr_00006|M.leprae_Br4923 PTAGLIVGSQGIADAYKTGRGSIRIRGVVEVEEDSRGRTSLVITELPYQV
MAB_0019|M.abscessus_ATCC_1997 PTSGLIVGTQGIHDAYTTGRGSIRMRGVAEIEEDSKGRTSLVITELPYQV
TH_0756|M.thermoresistible__bu PTAGLIVGSQGIHDAYTTGRGSIKMRGVVDIEEDSRGRTSLVITELPYQV
MSMEG_0456|M.smegmatis_MC2_155 GSGASVVGTAGLREYITTGRGHFTVRGTISVE-----GKNCIITELPPGV
Mvan_5898|M.vanbaalenii_PYR-1 GCGATVVGETGLREYITTGRGQFTVRGTVSVD-----GKNCIVTELPPGV
Mflv_0497|M.gilvum_PYR-GCK GCGASVIGTDGLREYITTGRGLFTVRGTVSVD-----GKNCIITELPPGV
.. ::* * : .**** : :**. .:: .. ::**** *
MMAR_0006|M.marinum_M NHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVVEIKRDAVAKVVL
MUL_0006|M.ulcerans_Agy99 NHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVVEIKRDAVAKVVL
Mb0006|M.bovis_AF2122/97 NHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVIEIKRDAVAKVVI
Rv0006|M.tuberculosis_H37Rv NHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVIEIKRDAVAKVVI
MAV_0006|M.avium_104 NHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVIELKRDAVAKVVL
MLBr_00006|M.leprae_Br4923 NHDNFITSIAEQVRTGRLAGISNVEDQGSDRVGVRIVIEIKRDAVAKVVL
MAB_0019|M.abscessus_ATCC_1997 NHDNFITSIAEQVRDGKIAGISNIEDQSSDRVGLRIVVVLKRDAVAKVVL
TH_0756|M.thermoresistible__bu NHDNFIASIAEQVRDGKIAGIANVEDQSSDRVGLRMVIELKRDAVAKVVL
MSMEG_0456|M.smegmatis_MC2_155 ASNTVQDRIRALVESGEMSGVADMSDLTDRRNGLRIVVTAKRGHNAEQIR
Mvan_5898|M.vanbaalenii_PYR-1 ASNTVQDRIRALVESGELSGVADMSDLTDRRNGLRIVVTAKRGHSAEQIR
Mflv_0497|M.gilvum_PYR-GCK ASGTVQDRIRALVESGEMAGVADMSDLTDRRNGLRIVVTAKRGHSAEQIR
... * *. *.::*::::.* . * *:*:*: **. *: :
MMAR_0006|M.marinum_M NNLYKHTQLQTSFGANMLSIVDG-VPRTLRLDQMIRYYVEHQLDVIIRRT
MUL_0006|M.ulcerans_Agy99 NNLYKHTQLQTSFGANMLSIVDG-VPRTLRLDQMIRYYVEHQLDVTIRRT
Mb0006|M.bovis_AF2122/97 NNLYKHTQLQTSFGANMLAIVDG-VPRTLRLDQLIRYYVDHQLDVIVRRT
Rv0006|M.tuberculosis_H37Rv NNLYKHTQLQTSFGANMLAIVDG-VPRTLRLDQLIRYYVDHQLDVIVRRT
MAV_0006|M.avium_104 NNLYKHTQLQTSFGANMLAIVDG-VPRTLRLDQLIRHYVDHQLDVIVRRT
MLBr_00006|M.leprae_Br4923 NNLYKHTQLQTSFGANMLSIVDG-VPRTLRLDQMICYYVEHQLDVIVRRT
MAB_0019|M.abscessus_ATCC_1997 NNLYKHTQLQTSFGANMLSIVDG-VPRTLRLDQLIRLYVNHQLDVIIRRT
TH_0756|M.thermoresistible__bu NNLYKHTQLQTSFGVNMLAIVDG-VPRTLRLDQMIRLYTDHQIDVIVRRT
MSMEG_0456|M.smegmatis_MC2_155 DQLLALTPLESTFAASLVALDENRVPRWWSVRDLIMAFLQLRDSVVLHRS
Mvan_5898|M.vanbaalenii_PYR-1 EQLLALTPLESTFAASLVALDENRVPRWWSVRELISAFLHLRDSVVVRRS
Mflv_0497|M.gilvum_PYR-GCK DQLLALTPLESSFAASLVALDENRVPRWWSVRELIAAFLSLRDSVVLHRS
::* * *:::*...:::: :. *** : ::* : : .* ::*:
MMAR_0006|M.marinum_M TYRLRKANERAHILRGLVKALDALDEVIALIRASQTVDIARVGLIELLDI
MUL_0006|M.ulcerans_Agy99 TYRLRKANERAHILRGLVKALDALDEVIALIRASQTVDIARAGLIELLGI
Mb0006|M.bovis_AF2122/97 TYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARAGLIELLDI
Rv0006|M.tuberculosis_H37Rv TYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARAGLIELLDI
MAV_0006|M.avium_104 TYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARQGLIELLDI
MLBr_00006|M.leprae_Br4923 TYRLRKANERAHILRGLVKALDALDEVITLIRASQTVDIARVGVVELLDI
MAB_0019|M.abscessus_ATCC_1997 RYRLRKANERAHILRGLVKALDALDEVIALIRASQTVDIARTGLIELLDV
TH_0756|M.thermoresistible__bu RYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARDGLMELLEI
MSMEG_0456|M.smegmatis_MC2_155 EYRLEKVTARRHLVAGLMKIHLDIDAAVAVIRGSETVDEARKGLQERFKI
Mvan_5898|M.vanbaalenii_PYR-1 EYRLEKVTARRHLVAGLMTIHLDIDAAVAVIRGSETVDEARQGLQERFGI
Mflv_0497|M.gilvum_PYR-GCK EYRLEKVTARRHLVAGLMTIHLDIDTAVAIIRASDTVDDARLGLQERFAI
***.*.. * *:: **:. :* .:::**.*:*** ** *: * : :
MMAR_0006|M.marinum_M DEIQAQAILDMQLRRLAALERQRIVDDLAKIEAEIADLEDILAKPERQRA
MUL_0006|M.ulcerans_Agy99 DEIQAQAILDMQLRRLAALERERIVDDLAKIEAEIADLEDILAKPERQRA
Mb0006|M.bovis_AF2122/97 DEIQAQAILDMQLRRLAALERQRIIDDLAKIEAEIADLEDILAKPERQRG
Rv0006|M.tuberculosis_H37Rv DEIQAQAILDMQLRRLAALERQRIIDDLAKIEAEIADLEDILAKPERQRG
MAV_0006|M.avium_104 DEIQAQAILDMQLRRLAALERQRIIDDLAKIEAEIADLEDILAKPERQRG
MLBr_00006|M.leprae_Br4923 DDIQAQAILDMQLRRLAALERQRIIDDLAKIEVEIADLGDILAKPERRRG
MAB_0019|M.abscessus_ATCC_1997 DEIQAQAILDMQLRRLAALERQKIIDDLAKIEAEIADLEDILAKPERQRA
TH_0756|M.thermoresistible__bu DEIQAQAILDMQLRRLAALERQRIVDDLAKIEAEIADLEDILAKPERQRG
MSMEG_0456|M.smegmatis_MC2_155 DAEQADYVLSLQLRRLTKLDVIELQAEAEKLDAEFAELNDLVTNPESRRK
Mvan_5898|M.vanbaalenii_PYR-1 DAEQADYVLALQLRRLTKLDVIELQAEAEKLDAEFAELTELVANPDARRK
Mflv_0497|M.gilvum_PYR-GCK DAVQADYVLALQLRRLTKLDVIELQAEAEKLDAEFAALTELVSNPDARRK
* **: :* :*****: *: .: : *::.*:* * :::::*: :*
MMAR_0006|M.marinum_M IVRDELAEIVDKHGDDRRTRIIAADGEVSDEDLIAREDVVVTITETGYAK
MUL_0006|M.ulcerans_Agy99 IVRDELAEIVDKHGDDRRTRIIAADGDVSDEDLIAREDVVVTITETGYAK
Mb0006|M.bovis_AF2122/97 IVRDELAEIVDRHGDDRRTRIIAADGDVSDEDLIAREDVVVTITETGYAK
Rv0006|M.tuberculosis_H37Rv IVRDELAEIVDRHGDDRRTRIIAADGDVSDEDLIAREDVVVTITETGYAK
MAV_0006|M.avium_104 IVRDELAEIVEKHGDARRTRIVAADGDVSDEDLIAREDVVVTITETGYAK
MLBr_00006|M.leprae_Br4923 IIRNELTEIAEKYGDDRRTRIIAVDGDVNDEDLIAREEVVVTITETGYAK
MAB_0019|M.abscessus_ATCC_1997 IVKDELAEITEKYGDDRRTRIISADGDVADEDLIAREDVVVTITETGYAK
TH_0756|M.thermoresistible__bu IVRDELKEIVDKHGDDRRSRLIAADGEVSDEDLIAREDVVVTITETGYAK
MSMEG_0456|M.smegmatis_MC2_155 VIDKELVETAKLFKGPEYDRRTVLDFDATP--------VSRG-DEDGSRE
Mvan_5898|M.vanbaalenii_PYR-1 VIDEELVETAKLFKGPEFDRRTVLDFDATP--------VSAGADEDGPRE
Mflv_0497|M.gilvum_PYR-GCK VIDQELVETAKLFKGPEYDRRTVLDYDATP--------VAANNSEDGPRE
:: .** * .. . . . * * :. * * * :
MMAR_0006|M.marinum_M RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
MUL_0006|M.ulcerans_Agy99 RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
Mb0006|M.bovis_AF2122/97 RTKTDLYRSQKRGGKGVQGAGLKQDDIVAHFFVCSTHDLILFFTTQGRVY
Rv0006|M.tuberculosis_H37Rv RTKTDLYRSQKRGGKGVQGAGLKQDDIVAHFFVCSTHDLILFFTTQGRVY
MAV_0006|M.avium_104 RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
MLBr_00006|M.leprae_Br4923 RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
MAB_0019|M.abscessus_ATCC_1997 RTKTDLYRSQKRGGKGVQGAGLKQDDIVKHFFVCSTHDWILFFTTKGRVY
TH_0756|M.thermoresistible__bu RTKTDLYRTQKRGGKGVQGAGLKQDDIVNHFFVCSTHDWILFFTTQGRVY
MSMEG_0456|M.smegmatis_MC2_155 RKVNTAWRLDDRG---------------------------VFSDSHG---
Mvan_5898|M.vanbaalenii_PYR-1 RKVNAAWRLDDRG---------------------------VFSDSHG---
Mflv_0497|M.gilvum_PYR-GCK RKVNAAWRLDDRG---------------------------VFSDSHG---
*. . :* :.** :* ::*
MMAR_0006|M.marinum_M RAKAYELPEALRTARGQHVANLLAFQPEERIAQVIQIKSYEDAPYLVLAT
MUL_0006|M.ulcerans_Agy99 RAKAYELPEALRTARGQHVANLLAFQPEERIAQVIQIKSYEDAPYLVLAT
Mb0006|M.bovis_AF2122/97 RAKAYDLPEASRTARGQHVANLLAFQPEERIAQVIQIRGYTDAPYLVLAT
Rv0006|M.tuberculosis_H37Rv RAKAYDLPEASRTARGQHVANLLAFQPEERIAQVIQIRGYTDAPYLVLAT
MAV_0006|M.avium_104 RAKAYELPEASRTARGQHVANLLAFQPEERIAQVIQIRSYEDAPYLVLAT
MLBr_00006|M.leprae_Br4923 RAKAYELPEASRTARGQHVANLLAFQPEERIAQVIQIRSYEDAPYLVLAT
MAB_0019|M.abscessus_ATCC_1997 RAKAYDLPEAARTARGQHVANLLAFQPEERIAQVIQIKSYEDAPYLVLAT
TH_0756|M.thermoresistible__bu RAKAYDLPEMSRTARGQHVANLLAFQPNERIAQVIQIRSYEDAPYLVLAT
MSMEG_0456|M.smegmatis_MC2_155 ----------------------------ELLTSGLGWAVWSDG-RIKFTT
Mvan_5898|M.vanbaalenii_PYR-1 ----------------------------DLLTSGLGWAVWTDG-RVKFTT
Mflv_0497|M.gilvum_PYR-GCK ----------------------------TLLSSGLGWAVWTDG-RIKFTT
::. : : *. : ::*
MMAR_0006|M.marinum_M QNGLVKKSKLTDFDSNRSGGIVAINLRGEDELVGAVLCSSEEDLLLVSAN
MUL_0006|M.ulcerans_Agy99 QNGVVKKSKLTDFDSNRSGGIVAINLRGEDELVGAVLCSSEEDLLLVSAN
Mb0006|M.bovis_AF2122/97 RNGLVKKSKLTAFDSNRSGGIVAVNLRDNDELVGAVLCSADDDLLLVSAN
Rv0006|M.tuberculosis_H37Rv RNGLVKKSKLTDFDSNRSGGIVAVNLRDNDELVGAVLCSAGDDLLLVSAN
MAV_0006|M.avium_104 RNGLVKKTKLTDFDSNRSGGIVAINLRDNDELVGAVLCSAEDDLLLVSAN
MLBr_00006|M.leprae_Br4923 RAGLVKKSKLTDFDSNRSGGIVAINLRDNDELVGAVLCAADGDLLLVSAN
MAB_0019|M.abscessus_ATCC_1997 KNGLVKKSKLTEFDSNRSGGLVAVNLRDGDELVGAVLCSAEDDLLLVSAH
TH_0756|M.thermoresistible__bu RNGLVKKSRLSDFDSNRSGGIVAINLREGDEVVGAVLCSAEDDLLLVSAN
MSMEG_0456|M.smegmatis_MC2_155 GNGLPFKIRDIPVAPDITG------------LVRSGVLPEGYHLALVTRR
Mvan_5898|M.vanbaalenii_PYR-1 GNGLPFKIRDIPVAPDITG------------LLCSGVLAPGHHLALVTRR
Mflv_0497|M.gilvum_PYR-GCK GGGLPFKIRDIPVSPDITG------------LLQSGVLAPGSHLALITRR
*: * : . .: :* :: : : . .* *:: .
MMAR_0006|M.marinum_M GQSIRFSATDEALRPMGRATSGVQGMRFNADD----RLLSLNVVREGTYL
MUL_0006|M.ulcerans_Agy99 GQSIRFSATDEALRPMGRATSGVQGMRFNADD----RLLSLNVVREGTYL
Mb0006|M.bovis_AF2122/97 GQSIRFSATDEALRPMGRATSGVQGMRFNIDD----RLLSLNVVREGTYL
Rv0006|M.tuberculosis_H37Rv GQSIRFSATDEALRPMGRATSGVQGMRFNIDD----RLLSLNVVREGTYL
MAV_0006|M.avium_104 GQSIRFSATDEALRPMGRATSGVQGMRFNADD----YLLSLNVVREGTYL
MLBr_00006|M.leprae_Br4923 GQSIRFSATDEALRPMGRATSGVQGMRFNADD----RLLSLNVVREDTYL
MAB_0019|M.abscessus_ATCC_1997 GQSIRFSATDEALRPMGRATSGVQGMRFNGED----DLLSLNVVREGTYL
TH_0756|M.thermoresistible__bu GQSIRFSATDEALRPMGRATSGVQGMRFNEDD----RLLSLNVVRPDTYL
MSMEG_0456|M.smegmatis_MC2_155 GKILRVDP--ASVNPQGVAGNGVAGVKLAADG--DEVIAALPVSCANGEA
Mvan_5898|M.vanbaalenii_PYR-1 GKILRIDP--AAVNPQGAAGNGVAGVKLAAGDPEDAVIAALPVSCANGEA
Mflv_0497|M.gilvum_PYR-GCK GRVLRIDP--AAVNPQGAAGNGVAGVKLVADDPEDRVIAALPVSCANGEA
*: :*... ::.* * * .** *::: . : :* * .
MMAR_0006|M.marinum_M LVATAGGYAKRTGIEEYPVQGRGGKGVLTIMYDRRRGRLVGALIVDDDSE
MUL_0006|M.ulcerans_Agy99 LAATAGGYAKRTGIEEYPVQGRGGKGVLTIMYDRRRGRLVGALIVDDDSE
Mb0006|M.bovis_AF2122/97 LVATSGGYAKRTAIEEYPVQGRGGKGVLTVMYDRRRGRLVGALIVDDDSE
Rv0006|M.tuberculosis_H37Rv LVATSGGYAKRTAIEEYPVQGRGGKGVLTVMYDRRRGRLVGALIVDDDSE
MAV_0006|M.avium_104 LVATSGGYAKRTAIEEYPVQGRGGKGVLTVMYDRRRGRLVGALIVDEDSE
MLBr_00006|M.leprae_Br4923 LVATSGGYAKRTSIEEYPMQGRGGKGVLTVMYDRRRGSLVGAIVVDEDSE
MAB_0019|M.abscessus_ATCC_1997 LVATSGGYSKRTAIEEYPVQGRGGKGVLTVQYDPRRGSLVGALVVDEESE
TH_0756|M.thermoresistible__bu LVATSGGYAKRTPIEEYPVQNRGGKGVLTIQYDRRRGNLVGALVVDDETE
MSMEG_0456|M.smegmatis_MC2_155 ILSIAEKSWKVTEVADIPVKGRGGAGVAFHPFVK-----------GEDAL
Mvan_5898|M.vanbaalenii_PYR-1 LLSISEKSWKVTEVADIPVKGRGGAGVGFHPFVK-----------GEDAV
Mflv_0497|M.gilvum_PYR-GCK ILSISEKSWKVTEVADIPVKGRGGAGVGFHPFGK-----------GEDAL
: : : * * : : *::.*** ** : .:::
MMAR_0006|M.marinum_M LYAITSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEENA
MUL_0006|M.ulcerans_Agy99 LYAITSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEENA
Mb0006|M.bovis_AF2122/97 LYAVTSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEESG
Rv0006|M.tuberculosis_H37Rv LYAVTSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEESG
MAV_0006|M.avium_104 LYAITSGGGVIRTAAGQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEEAA
MLBr_00006|M.leprae_Br4923 LYAITSGGGVIRTTARQVRQAGRQTKGVRLMNLGEGDTLLAIARNAEESA
MAB_0019|M.abscessus_ATCC_1997 LYAITSGGGVIRTIAKQVRKAGRQTKGVRLMNLGEGDTLLAIAHNADEGD
TH_0756|M.thermoresistible__bu LYAITSSGGVIRTTARQIRKAGRQTKGVRLMNLAEGDTLVAIARNAEAGD
MSMEG_0456|M.smegmatis_MC2_155 LAASISKTGYVR-----GKRAVRPENRAKASIKGSGADVTPAPAE-----
Mvan_5898|M.vanbaalenii_PYR-1 LAATISATGYVR-----GKKAVRADNRAKASIKGSGVDVTPAG-------
Mflv_0497|M.gilvum_PYR-GCK LSAAISATGYVR-----GTRTVRAEKRAKASIKGSGADVTPADAAPAD--
* * * * :* :: * : .: ..* : .
MMAR_0006|M.marinum_M DEAVAEATDTDESSN
MUL_0006|M.ulcerans_Agy99 DEAVAEATDTDESSN
Mb0006|M.bovis_AF2122/97 DDNAVDANGADQTGN
Rv0006|M.tuberculosis_H37Rv DDNAVDANGADQTGN
MAV_0006|M.avium_104 DEAVEESDGAAGSDG
MLBr_00006|M.leprae_Br4923 DGGVG----------
MAB_0019|M.abscessus_ATCC_1997 ADPDEDAAGTTAGE-
TH_0756|M.thermoresistible__bu STDEVNTDPDSV---
MSMEG_0456|M.smegmatis_MC2_155 ---------------
Mvan_5898|M.vanbaalenii_PYR-1 ---------------
Mflv_0497|M.gilvum_PYR-GCK ---------------