For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDITLPPGDGSIQRVEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGLKPVHRRVLYAMLDSGFRPDRS HAKSARSVAETMGNYHPHGDASIYDTLVRMAQPWSLRYPLVDGQGNFGSPGNDPPAAMRYCVSGNSLVRL LFGKSIRIGDIVTGAQFNSDNPIDLKVLDRHGNPVVADYLFHSGEHQTYTVRTTEGYEITGTSNHPLLCL VNVGGIPTLLWKLIGEIRSGDYVVLQRIPPVEFGPADWYSTMEALLFGAFISGGFVFQDHAGFNSLDRDY FTMVVNAYDTVVGGLRCISSRITVSGSTLLELDVYNLIEFKKTRLSGLCGQRSADKLVPDWLWHSPSTVK RAFLQALFEGEGFSSILSRNIIEISYSTPSERLAADVQQMLLEFGVVSERYCHTVNEYKVVIANRAQVEM FFTQVGFGVTKQAKLIRDVVSMSPCVGMDINCVPGLATFIRKHCDNRWVEEDSFNQHNVDCVQHWHHHSA EIVGHIADPDIRAIVTDLTDGRFYYARVASVTDTGIQPVFSLHVDTEDHSFLTNGFISHNTEARLTPLAM EMLREIDEETVDFISNYDGRVQEPMVLPSRFPNLLANGSGGIAVGMATNIPPHNLYELADAVFWCLENHD ADEETMLVAVMERVKGPDFPTAGLIVGSQGIADAYKTGRGSIRIRGVVEVEEDSRGRTSLVITELPYQVN HDNFITSIAEQVRTGRLAGISNVEDQGSDRVGVRIVIEIKRDAVAKVVLNNLYKHTQLQTSFGANMLSIV DGVPRTLRLDQMICYYVEHQLDVIVRRTTYRLRKANERAHILRGLVKALDALDEVITLIRASQTVDIARV GVVELLDIDDIQAQAILDMQLRRLAALERQRIIDDLAKIEVEIADLGDILAKPERRRGIIRNELTEIAEK YGDDRRTRIIAVDGDVNDEDLIAREEVVVTITETGYAKRTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFF VCSTHDWILFFTTQGRVYRAKAYELPEASRTARGQHVANLLAFQPEERIAQVIQIRSYEDAPYLVLATRA GLVKKSKLTDFDSNRSGGIVAINLRDNDELVGAVLCAADGDLLLVSANGQSIRFSATDEALRPMGRATSG VQGMRFNADDRLLSLNVVREDTYLLVATSGGYAKRTSIEEYPMQGRGGKGVLTVMYDRRRGSLVGAIVVD EDSELYAITSGGGVIRTTARQVRQAGRQTKGVRLMNLGEGDTLLAIARNAEESADGGVG
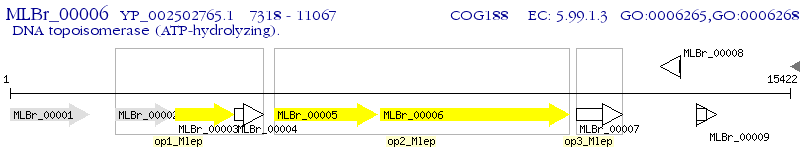
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. leprae Br4923 | MLBr_00006 | gyrA | - | 100% (1249) | putative DNA gyrase subunit A |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0006 | gyrA | 0.0 | 91.22% (695) | DNA gyrase subunit A |
| M. gilvum PYR-GCK | Mflv_0821 | - | 0.0 | 79.81% (1243) | DNA gyrase, A subunit |
| M. tuberculosis H37Rv | Rv0006 | gyrA | 0.0 | 91.22% (695) | DNA gyrase subunit A |
| M. abscessus ATCC 19977 | MAB_0019 | - | 0.0 | 86.99% (692) | DNA gyrase subunit A |
| M. marinum M | MMAR_0006 | gyrA | 0.0 | 90.83% (698) | DNA gyrase (subunit A) GyrA |
| M. avium 104 | MAV_0006 | gyrA | 0.0 | 91.12% (698) | DNA gyrase subunit A |
| M. smegmatis MC2 155 | MSMEG_0006 | gyrA | 0.0 | 88.15% (692) | DNA gyrase subunit A |
| M. thermoresistible (build 8) | TH_0756 | gyrA | 0.0 | 87.41% (691) | DNA GYRASE (SUBUNIT A) GYRA (DNA TOPOISOMERASE |
| M. ulcerans Agy99 | MUL_0006 | gyrA | 0.0 | 89.97% (698) | DNA gyrase subunit A |
| M. vanbaalenii PYR-1 | Mvan_0007 | - | 0.0 | 80.93% (1243) | DNA gyrase, A subunit |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0006|M.bovis_AF2122/97 MTDTTLPPDD--SLDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
Rv0006|M.tuberculosis_H37Rv MTDTTLPPDD--SLDRIEPVDIEQEMQRSYIDYAMSVIVGRALPEVRDGL
MLBr_00006|M.leprae_Br4923 MTDITLPPGDG-SIQRVEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MAV_0006|M.avium_104 MTDTTLPPGGD-AADRVEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MMAR_0006|M.marinum_M MTDTTLPPGGE-ASDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MUL_0006|M.ulcerans_Agy99 MTDTMLPPGGE-ASDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MAB_0019|M.abscessus_ATCC_1997 MTDTTLPPGGDDAVDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
Mflv_0821|M.gilvum_PYR-GCK MTDTTLPPGDE-AGDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
Mvan_0007|M.vanbaalenii_PYR-1 MTDTTLPPGGE-AGDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
TH_0756|M.thermoresistible__bu MTDTTLPPGGE-AGDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
MSMEG_0006|M.smegmatis_MC2_155 MTDTTLPPEGE-AHDRIEPVDIQQEMQRSYIDYAMSVIVGRALPEVRDGL
*** *** . : :*:*****:***************************
Mb0006|M.bovis_AF2122/97 KPVHRRVLYAMFDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
Rv0006|M.tuberculosis_H37Rv KPVHRRVLYAMFDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDSLVR
MLBr_00006|M.leprae_Br4923 KPVHRRVLYAMLDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MAV_0006|M.avium_104 KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MMAR_0006|M.marinum_M KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MUL_0006|M.ulcerans_Agy99 KPVHRRVLYAMYDSGFRPDHSHAKSARSVAETMGNYHPHGDASIYDTLVR
MAB_0019|M.abscessus_ATCC_1997 KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
Mflv_0821|M.gilvum_PYR-GCK KPVHRRVLYAMFDSGFRPDRGHAKSARSVAETMGNYHPHGDSSIYDTLVR
Mvan_0007|M.vanbaalenii_PYR-1 KPVHRRVLYAMFDSGFRPDRGHAKSARSVAETMGNYHPHGDASIYDTLVR
TH_0756|M.thermoresistible__bu KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
MSMEG_0006|M.smegmatis_MC2_155 KPVHRRVLYAMYDSGFRPDRSHAKSARSVAETMGNYHPHGDASIYDTLVR
*********** *******:.********************:****:***
Mb0006|M.bovis_AF2122/97 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
Rv0006|M.tuberculosis_H37Rv MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MLBr_00006|M.leprae_Br4923 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRYCVSGNSLVRLLFGKSIRIG
MAV_0006|M.avium_104 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MMAR_0006|M.marinum_M MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MUL_0006|M.ulcerans_Agy99 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MAB_0019|M.abscessus_ATCC_1997 MAQPWSLRYPLVDGQGNFGSPGNDPAAAMRY-------------------
Mflv_0821|M.gilvum_PYR-GCK MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRYCVTGDAMVRLPFGQSVRIA
Mvan_0007|M.vanbaalenii_PYR-1 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRYCLTGDSMVRLPFGQSVRIG
TH_0756|M.thermoresistible__bu MAQPWSMRYPLVDGQGNFGSPGNDPPAAMRY-------------------
MSMEG_0006|M.smegmatis_MC2_155 MAQPWSLRYPLVDGQGNFGSPGNDPPAAMRY-------------------
******:******************.*****
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 DIVTGAQFNSDNPIDLKVLDRHGNPVVADYLFHSGEHQTYTVRTTEGYEI
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK DIVKGARPNSDNDIDVKVLDRHGDPVVANALFHSGDHETFAVRTAEGYEV
Mvan_0007|M.vanbaalenii_PYR-1 DVVSGARPNSDNEIELKVLDRHGNPVLADALFHSGDHETFTVRTAEGYEV
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 TGTSNHPLLCLVNVGGIPTLLWKLIGEIRSGDYVVLQRIPPVEFGPADWY
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK TGTSNHPLLCLVDVGGVPTLLWKLIGEIRPGDHVVLQRTPPVEFGPGDWR
Mvan_0007|M.vanbaalenii_PYR-1 TGTANHPLLCLVDVGGVPTLLWKLIAEIRPSDRVVLQRTPPVEFGPADWY
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 STMEALLFGAFISGGFVFQDHAGFNSLDRDYFTMVVNAYDTVVGGLRCIS
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK EVMEALLLGAFISDGFVSESRAGFNNLDRDYFNMVLAAYDAIVGGPRYVY
Mvan_0007|M.vanbaalenii_PYR-1 ETMEALLLGAFISEGFVSESRAGFNNLDRDYFNMVVAAYDTVVGGPRYVY
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 SRITVSGSTLLELDVYNLIEFKKTRLSGLCGQRSADKLVPDWLWHSPSTV
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK ERRIASGSKLLELDVHNLRALDSSRLAELKSQRSAAKTVPNWLWNSTAAA
Mvan_0007|M.vanbaalenii_PYR-1 ERKIASGSTLLELDIQNLASLRQSRLVGLLGQRSAAKMVPEWLWNSAAAV
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 KRAFLQALFEGEGFSSILSRNIIEISYSTPSERLAADVQQMLLEFGVVSE
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK KKAFLQALFEGDGSCSALPHNTIQLSYSTRSGQLAKDVQNMLLEFGVISR
Mvan_0007|M.vanbaalenii_PYR-1 KRAFLQALFEGDGSCSALPRNTIQVSYSTRSGQLAKDVQQMLLEFGVISR
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 RYCHTVNEYKVVIANRAQVEMFFTQVGFGVTKQAKLIRDVVSMSP-CVGM
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK RYLHATGEHKVVITNRAHAEDFCLRIGFGGAKQEKLQELLAALPARAAGL
Mvan_0007|M.vanbaalenii_PYR-1 RYLHATGEHKVVITNRSSAEAFCTRVGFGGAKQHKLQKLLSDLPERAAGL
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 DINCVPGLATFIRKHCDNRWVEEDSFNQHNVDCVQHWHHHSAEIVGHIAD
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK DGDHVPGLATFIRSRSGGRWIDKEWLRKHNIDRLTRWRRDREEILSHISD
Mvan_0007|M.vanbaalenii_PYR-1 DGDHVPGLAAFIRRESGSKWVDKEWLRKHNIDRLSRWRRDGSEILSRIAD
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --------------------------------------------------
Rv0006|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00006|M.leprae_Br4923 PDIRAIVTDLTDGRFYYARVASVTDTGIQPVFSLHVDTEDHSFLTNGFIS
MAV_0006|M.avium_104 --------------------------------------------------
MMAR_0006|M.marinum_M --------------------------------------------------
MUL_0006|M.ulcerans_Agy99 --------------------------------------------------
MAB_0019|M.abscessus_ATCC_1997 --------------------------------------------------
Mflv_0821|M.gilvum_PYR-GCK NDVRSIATELTDGRFYYARVASVSDAGVQPVYSLRVDTDDHSFITNGFVS
Mvan_0007|M.vanbaalenii_PYR-1 PDVRAIATELTDGRFYYAEVATVVSAGVQPVYSLRVDTDDHSFISNGFVS
TH_0756|M.thermoresistible__bu --------------------------------------------------
MSMEG_0006|M.smegmatis_MC2_155 --------------------------------------------------
Mb0006|M.bovis_AF2122/97 --TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANG
Rv0006|M.tuberculosis_H37Rv --TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANG
MLBr_00006|M.leprae_Br4923 HNTEARLTPLAMEMLREIDEETVDFISNYDGRVQEPMVLPSRFPNLLANG
MAV_0006|M.avium_104 --TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANG
MMAR_0006|M.marinum_M --TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANG
MUL_0006|M.ulcerans_Agy99 --TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANG
MAB_0019|M.abscessus_ATCC_1997 --TEARLTPLAMEMLREIDEETVDFIPNYDGRVMEPTVLPSRFPNLLANG
Mflv_0821|M.gilvum_PYR-GCK HNTEARLTPLAMEMLREIDEETVDFVPNYDGRVQEPTVLPSRFPNLLANG
Mvan_0007|M.vanbaalenii_PYR-1 HNTEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANG
TH_0756|M.thermoresistible__bu --TEARLTPLAMEMLREIDEETVDFVPNYDGRVQEPTVLPSRFPNLLANG
MSMEG_0006|M.smegmatis_MC2_155 --TEARLTPLAMEMLREIDEETVDFIPNYDGRVQEPTVLPSRFPNLLANG
***********************:.****** ** *************
Mb0006|M.bovis_AF2122/97 SGGIAVGMATNIPPHNLRELADAVFWALENHDADEEETLAAVMGRVKGPD
Rv0006|M.tuberculosis_H37Rv SGGIAVGMATNIPPHNLRELADAVFWALENHDADEEETLAAVMGRVKGPD
MLBr_00006|M.leprae_Br4923 SGGIAVGMATNIPPHNLYELADAVFWCLENHDADEETMLVAVMERVKGPD
MAV_0006|M.avium_104 SGGIAVGMATNIPPHNLGELAEAVFWALDNYEADEEATLAAVMERVKGPD
MMAR_0006|M.marinum_M SGGIAVGMATNIPPHNLRELAEAVFWCLENHDADEEATLAAVTERVKGPD
MUL_0006|M.ulcerans_Agy99 SGGIAVGMATNIPPHNLRELAEAVFWCLENHDADEEATLAAVTDRVKGPD
MAB_0019|M.abscessus_ATCC_1997 SGGIAVGMATNMPPHNLRELAEAVYWALDNHEADEETTLKAVCEKITGPD
Mflv_0821|M.gilvum_PYR-GCK SGGIAVGMATNMPPHNLRELAEAVYWCLDNHEADEEATLDAVCERVKGPD
Mvan_0007|M.vanbaalenii_PYR-1 SGGIAVGMATNMPPHNLRELAEAVYWCLENYEADEEATLEAVCERVKGPD
TH_0756|M.thermoresistible__bu SGGIAVGMATNIPPHNLRELAEAVYWCLENYEADEETTLAAVMERVKGPD
MSMEG_0006|M.smegmatis_MC2_155 SGGIAVGMATNIPPHNLGELAEAVYWCLENYEADEEATCEAVMERVKGPD
***********:***** ***:**:*.*:*::**** ** ::.***
Mb0006|M.bovis_AF2122/97 FPTAGLIVGSQGTADAYKTGRGSIRMRGVVEVEEDSRGRTSLVITELPYQ
Rv0006|M.tuberculosis_H37Rv FPTAGLIVGSQGTADAYKTGRGSIRMRGVVEVEEDSRGRTSLVITELPYQ
MLBr_00006|M.leprae_Br4923 FPTAGLIVGSQGIADAYKTGRGSIRIRGVVEVEEDSRGRTSLVITELPYQ
MAV_0006|M.avium_104 FPTSGLIVGTQGIADAYKTGRGSIRMRGVVEVEEDSRGRTSLVITELPYQ
MMAR_0006|M.marinum_M FPTSGLIVGTQGISDAYKTGRGSIRMRGVVEIEEDSRGRTSLVITELPYQ
MUL_0006|M.ulcerans_Agy99 FPTSGLIVGTQGISDAYKTGRGSIRMRGVVEIEEDSRGRTSLVITELPYQ
MAB_0019|M.abscessus_ATCC_1997 FPTSGLIVGTQGIHDAYTTGRGSIRMRGVAEIEEDSKGRTSLVITELPYQ
Mflv_0821|M.gilvum_PYR-GCK FPTYGLIVGSQGIDDTYKTGRGSIRMRGVVEIEEDARGRTSLVITELPYQ
Mvan_0007|M.vanbaalenii_PYR-1 FPTHGLIVGSQGISDTYTTGRGSIRMRGVVEIEEDSRGRTSLVITELPYQ
TH_0756|M.thermoresistible__bu FPTAGLIVGSQGIHDAYTTGRGSIKMRGVVDIEEDSRGRTSLVITELPYQ
MSMEG_0006|M.smegmatis_MC2_155 FPTSGLIVGTQGIEDTYKTGRGSIKMRGVVEIEEDSRGRTSIVITELPYQ
*** *****:** *:*.******::***.::***::****:********
Mb0006|M.bovis_AF2122/97 VNHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVIEIKRDAVAKVV
Rv0006|M.tuberculosis_H37Rv VNHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVIEIKRDAVAKVV
MLBr_00006|M.leprae_Br4923 VNHDNFITSIAEQVRTGRLAGISNVEDQGSDRVGVRIVIEIKRDAVAKVV
MAV_0006|M.avium_104 VNHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVIELKRDAVAKVV
MMAR_0006|M.marinum_M VNHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVVEIKRDAVAKVV
MUL_0006|M.ulcerans_Agy99 VNHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVVEIKRDAVAKVV
MAB_0019|M.abscessus_ATCC_1997 VNHDNFITSIAEQVRDGKIAGISNIEDQSSDRVGLRIVVVLKRDAVAKVV
Mflv_0821|M.gilvum_PYR-GCK VNHDNFITSIAEQVRDGKLTGISNIEDQSSDRVGLRIVVEIKRDAVAKVV
Mvan_0007|M.vanbaalenii_PYR-1 VNHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVVEIKRDAVAKVV
TH_0756|M.thermoresistible__bu VNHDNFIASIAEQVRDGKIAGIANVEDQSSDRVGLRMVIELKRDAVAKVV
MSMEG_0006|M.smegmatis_MC2_155 VNHDNFITSIAEQVRDGKLAGISNIEDQSSDRVGLRIVVELKRDAVAKVV
*******:******* *:::**:*:***.*****:*:*: :*********
Mb0006|M.bovis_AF2122/97 INNLYKHTQLQTSFGANMLAIVDGVPRTLRLDQLIRYYVDHQLDVIVRRT
Rv0006|M.tuberculosis_H37Rv INNLYKHTQLQTSFGANMLAIVDGVPRTLRLDQLIRYYVDHQLDVIVRRT
MLBr_00006|M.leprae_Br4923 LNNLYKHTQLQTSFGANMLSIVDGVPRTLRLDQMICYYVEHQLDVIVRRT
MAV_0006|M.avium_104 LNNLYKHTQLQTSFGANMLAIVDGVPRTLRLDQLIRHYVDHQLDVIVRRT
MMAR_0006|M.marinum_M LNNLYKHTQLQTSFGANMLSIVDGVPRTLRLDQMIRYYVEHQLDVIIRRT
MUL_0006|M.ulcerans_Agy99 LNNLYKHTQLQTSFGANMLSIVDGVPRTLRLDQMIRYYVEHQLDVTIRRT
MAB_0019|M.abscessus_ATCC_1997 LNNLYKHTQLQTSFGANMLSIVDGVPRTLRLDQLIRLYVNHQLDVIIRRT
Mflv_0821|M.gilvum_PYR-GCK LNNLYKHTQLQTSFGANMLSIVDGVPRTLRLDQLIRHYVNHQLDVIVRRT
Mvan_0007|M.vanbaalenii_PYR-1 LNNLYKHTQLQTSFGANMLSIVDGVPRTLRLDQLIRLYVNHQLDVIVRRT
TH_0756|M.thermoresistible__bu LNNLYKHTQLQTSFGVNMLAIVDGVPRTLRLDQMIRLYTDHQIDVIVRRT
MSMEG_0006|M.smegmatis_MC2_155 LNNLYKHTQLQTSFGANMLSIVDGVPRTLRLDQLIRLYVDHQLDVIVRRT
:**************.***:*************:* *.:**:** :***
Mb0006|M.bovis_AF2122/97 TYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARAGLIELLDI
Rv0006|M.tuberculosis_H37Rv TYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARAGLIELLDI
MLBr_00006|M.leprae_Br4923 TYRLRKANERAHILRGLVKALDALDEVITLIRASQTVDIARVGVVELLDI
MAV_0006|M.avium_104 TYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARQGLIELLDI
MMAR_0006|M.marinum_M TYRLRKANERAHILRGLVKALDALDEVIALIRASQTVDIARVGLIELLDI
MUL_0006|M.ulcerans_Agy99 TYRLRKANERAHILRGLVKALDALDEVIALIRASQTVDIARAGLIELLGI
MAB_0019|M.abscessus_ATCC_1997 RYRLRKANERAHILRGLVKALDALDEVIALIRASQTVDIARTGLIELLDV
Mflv_0821|M.gilvum_PYR-GCK RYRLRKANERAHILRGLVKALDALDEVIALIRASQTVEIARSGLIELLDI
Mvan_0007|M.vanbaalenii_PYR-1 TYRLRKANERAHILRGLVKALDALDEVIALIRASQTVEIARSGLIELLDI
TH_0756|M.thermoresistible__bu RYRLRKANERAHILRGLVKALDALDEVIALIRASETVDIARDGLMELLEI
MSMEG_0006|M.smegmatis_MC2_155 RYRLRKANERAHILRGLVKALDALDEVIALIRASQTVDIARAGLIELLDI
***************************:*****:**:*** *::*** :
Mb0006|M.bovis_AF2122/97 DEIQAQAILDMQLRRLAALERQRIIDDLAKIEAEIADLEDILAKPERQRG
Rv0006|M.tuberculosis_H37Rv DEIQAQAILDMQLRRLAALERQRIIDDLAKIEAEIADLEDILAKPERQRG
MLBr_00006|M.leprae_Br4923 DDIQAQAILDMQLRRLAALERQRIIDDLAKIEVEIADLGDILAKPERRRG
MAV_0006|M.avium_104 DEIQAQAILDMQLRRLAALERQRIIDDLAKIEAEIADLEDILAKPERQRG
MMAR_0006|M.marinum_M DEIQAQAILDMQLRRLAALERQRIVDDLAKIEAEIADLEDILAKPERQRA
MUL_0006|M.ulcerans_Agy99 DEIQAQAILDMQLRRLAALERERIVDDLAKIEAEIADLEDILAKPERQRA
MAB_0019|M.abscessus_ATCC_1997 DEIQAQAILDMQLRRLAALERQKIIDDLAKIEAEIADLEDILAKPERQRA
Mflv_0821|M.gilvum_PYR-GCK DEIQAQAILDMQLRRLAALERQRIVDDLAKIEAEIADLEDILAKPERQRA
Mvan_0007|M.vanbaalenii_PYR-1 DEIQAQAILDMQLRRLAALERQRIIDDLAKIEAEIADLEDILAKPERQRA
TH_0756|M.thermoresistible__bu DEIQAQAILDMQLRRLAALERQRIVDDLAKIEAEIADLEDILAKPERQRG
MSMEG_0006|M.smegmatis_MC2_155 DDIQAQAILDMQLRRLAALERQKIVDDLAKIEAEIADLEDILAKPERQRG
*:*******************::*:*******.***** ********:*.
Mb0006|M.bovis_AF2122/97 IVRDELAEIVDRHGDDRRTRIIAADGDVSDEDLIAREDVVVTITETGYAK
Rv0006|M.tuberculosis_H37Rv IVRDELAEIVDRHGDDRRTRIIAADGDVSDEDLIAREDVVVTITETGYAK
MLBr_00006|M.leprae_Br4923 IIRNELTEIAEKYGDDRRTRIIAVDGDVNDEDLIAREEVVVTITETGYAK
MAV_0006|M.avium_104 IVRDELAEIVEKHGDARRTRIVAADGDVSDEDLIAREDVVVTITETGYAK
MMAR_0006|M.marinum_M IVRDELAEIVDKHGDDRRTRIIAADGEVSDEDLIAREDVVVTITETGYAK
MUL_0006|M.ulcerans_Agy99 IVRDELAEIVDKHGDDRRTRIIAADGDVSDEDLIAREDVVVTITETGYAK
MAB_0019|M.abscessus_ATCC_1997 IVKDELAEITEKYGDDRRTRIISADGDVADEDLIAREDVVVTITETGYAK
Mflv_0821|M.gilvum_PYR-GCK IVHDELKEIVDKYGDDRRTRIIPADGDVADEDLIAREEVVVTITETGYAK
Mvan_0007|M.vanbaalenii_PYR-1 IVHDELKELVDKYGDDRRTRIIPADGDVADEDLIAREEVVVTITETGYAK
TH_0756|M.thermoresistible__bu IVRDELKEIVDKHGDDRRSRLIAADGEVSDEDLIAREDVVVTITETGYAK
MSMEG_0006|M.smegmatis_MC2_155 IVRDELKEIVDKHGDARRTRIVPADGEVSDEDLIAREDVVVTITETGYAK
*:::** *:.:::** **:*::..**:* ********:************
Mb0006|M.bovis_AF2122/97 RTKTDLYRSQKRGGKGVQGAGLKQDDIVAHFFVCSTHDLILFFTTQGRVY
Rv0006|M.tuberculosis_H37Rv RTKTDLYRSQKRGGKGVQGAGLKQDDIVAHFFVCSTHDLILFFTTQGRVY
MLBr_00006|M.leprae_Br4923 RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
MAV_0006|M.avium_104 RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
MMAR_0006|M.marinum_M RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
MUL_0006|M.ulcerans_Agy99 RTKTDLYRSQKRGGKGVQGAGLKQDDIVRHFFVCSTHDWILFFTTQGRVY
MAB_0019|M.abscessus_ATCC_1997 RTKTDLYRSQKRGGKGVQGAGLKQDDIVKHFFVCSTHDWILFFTTKGRVY
Mflv_0821|M.gilvum_PYR-GCK RTKSDLYRSQKRGGKGVQGAGLKQDDIVNHFFVCSTHDWILFFTTQGRVY
Mvan_0007|M.vanbaalenii_PYR-1 RTKSDLYRSQKRGGKGVQGAGLKQDDIVNHFFVCSTHDWILFFTTQGRVY
TH_0756|M.thermoresistible__bu RTKTDLYRTQKRGGKGVQGAGLKQDDIVNHFFVCSTHDWILFFTTQGRVY
MSMEG_0006|M.smegmatis_MC2_155 RTKTDLYRSQKRGGKGVQGAGLKQDDMVNHFFVCSTHDWILFFTTQGRVY
***:****:*****************:* ********* ******:****
Mb0006|M.bovis_AF2122/97 RAKAYDLPEASRTARGQHVANLLAFQPEERIAQVIQIRGYTDAPYLVLAT
Rv0006|M.tuberculosis_H37Rv RAKAYDLPEASRTARGQHVANLLAFQPEERIAQVIQIRGYTDAPYLVLAT
MLBr_00006|M.leprae_Br4923 RAKAYELPEASRTARGQHVANLLAFQPEERIAQVIQIRSYEDAPYLVLAT
MAV_0006|M.avium_104 RAKAYELPEASRTARGQHVANLLAFQPEERIAQVIQIRSYEDAPYLVLAT
MMAR_0006|M.marinum_M RAKAYELPEALRTARGQHVANLLAFQPEERIAQVIQIKSYEDAPYLVLAT
MUL_0006|M.ulcerans_Agy99 RAKAYELPEALRTARGQHVANLLAFQPEERIAQVIQIKSYEDAPYLVLAT
MAB_0019|M.abscessus_ATCC_1997 RAKAYDLPEAARTARGQHVANLLAFQPEERIAQVIQIKSYEDAPYLVLAT
Mflv_0821|M.gilvum_PYR-GCK RAKAYDLPEASRTARGQHVANLLAFQPEERIAQVIQLKTYEDAPYLVLAT
Mvan_0007|M.vanbaalenii_PYR-1 RAKAYDLPEASRTARGQHVANLLAFQPEERIAQVIQLKSYEDAPYLVLAT
TH_0756|M.thermoresistible__bu RAKAYDLPEMSRTARGQHVANLLAFQPNERIAQVIQIRSYEDAPYLVLAT
MSMEG_0006|M.smegmatis_MC2_155 RAKAYELPEASRTARGQHVANLLAFQPEERIAQVIQIKSYEDAPYLVLAT
*****:*** ****************:********:: * *********
Mb0006|M.bovis_AF2122/97 RNGLVKKSKLTAFDSNRSGGIVAVNLRDNDELVGAVLCSADDDLLLVSAN
Rv0006|M.tuberculosis_H37Rv RNGLVKKSKLTDFDSNRSGGIVAVNLRDNDELVGAVLCSAGDDLLLVSAN
MLBr_00006|M.leprae_Br4923 RAGLVKKSKLTDFDSNRSGGIVAINLRDNDELVGAVLCAADGDLLLVSAN
MAV_0006|M.avium_104 RNGLVKKTKLTDFDSNRSGGIVAINLRDNDELVGAVLCSAEDDLLLVSAN
MMAR_0006|M.marinum_M QNGLVKKSKLTDFDSNRSGGIVAINLRGEDELVGAVLCSSEEDLLLVSAN
MUL_0006|M.ulcerans_Agy99 QNGVVKKSKLTDFDSNRSGGIVAINLRGEDELVGAVLCSSEEDLLLVSAN
MAB_0019|M.abscessus_ATCC_1997 KNGLVKKSKLTEFDSNRSGGLVAVNLRDGDELVGAVLCSAEDDLLLVSAH
Mflv_0821|M.gilvum_PYR-GCK RNGLVKKSRLVDFDSNRSGGIVAVNLRDGDELVGAVLCSADDDLLLVSAK
Mvan_0007|M.vanbaalenii_PYR-1 RNGLVKKSRLVDFDSNRSGGIVAVNLRDGDELVGAVLCSAEDDLLLVSAK
TH_0756|M.thermoresistible__bu RNGLVKKSRLSDFDSNRSGGIVAINLREGDEVVGAVLCSAEDDLLLVSAN
MSMEG_0006|M.smegmatis_MC2_155 RNGLVKKSKLSDFDSNRSGGIVAINLREGDELVGAVLCSAEDDLLLVSAN
: *:***::* ********:**:*** **:******:: *******:
Mb0006|M.bovis_AF2122/97 GQSIRFSATDEALRPMGRATSGVQGMRFNIDDRLLSLNVVREGTYLLVAT
Rv0006|M.tuberculosis_H37Rv GQSIRFSATDEALRPMGRATSGVQGMRFNIDDRLLSLNVVREGTYLLVAT
MLBr_00006|M.leprae_Br4923 GQSIRFSATDEALRPMGRATSGVQGMRFNADDRLLSLNVVREDTYLLVAT
MAV_0006|M.avium_104 GQSIRFSATDEALRPMGRATSGVQGMRFNADDYLLSLNVVREGTYLLVAT
MMAR_0006|M.marinum_M GQSIRFSATDEALRPMGRATSGVQGMRFNADDRLLSLNVVREGTYLLVAT
MUL_0006|M.ulcerans_Agy99 GQSIRFSATDEALRPMGRATSGVQGMRFNADDRLLSLNVVREGTYLLAAT
MAB_0019|M.abscessus_ATCC_1997 GQSIRFSATDEALRPMGRATSGVQGMRFNGEDDLLSLNVVREGTYLLVAT
Mflv_0821|M.gilvum_PYR-GCK GQSIRFSATDEALRPMGRATSGVQGMRFNEEDQLLSLNVVREGTYLLVAT
Mvan_0007|M.vanbaalenii_PYR-1 GQSIRFSATDEALRPMGRATSGVQGMRFNEEDQLLSLNVVREDTYLLVAT
TH_0756|M.thermoresistible__bu GQSIRFSATDEALRPMGRATSGVQGMRFNEDDRLLSLNVVRPDTYLLVAT
MSMEG_0006|M.smegmatis_MC2_155 GQSIRFSATDEALRPMGRATSGVQGMRFNEDDRLLSLNVVRPDTYLLVAT
***************************** :* ******** .****.**
Mb0006|M.bovis_AF2122/97 SGGYAKRTAIEEYPVQGRGGKGVLTVMYDRRRGRLVGALIVDDDSELYAV
Rv0006|M.tuberculosis_H37Rv SGGYAKRTAIEEYPVQGRGGKGVLTVMYDRRRGRLVGALIVDDDSELYAV
MLBr_00006|M.leprae_Br4923 SGGYAKRTSIEEYPMQGRGGKGVLTVMYDRRRGSLVGAIVVDEDSELYAI
MAV_0006|M.avium_104 SGGYAKRTAIEEYPVQGRGGKGVLTVMYDRRRGRLVGALIVDEDSELYAI
MMAR_0006|M.marinum_M AGGYAKRTGIEEYPVQGRGGKGVLTIMYDRRRGRLVGALIVDDDSELYAI
MUL_0006|M.ulcerans_Agy99 AGGYAKRTGIEEYPVQGRGGKGVLTIMYDRRRGRLVGALIVDDDSELYAI
MAB_0019|M.abscessus_ATCC_1997 SGGYSKRTAIEEYPVQGRGGKGVLTVQYDPRRGSLVGALVVDEESELYAI
Mflv_0821|M.gilvum_PYR-GCK SGGYAKRTAIEEYSAQGRGGKGILTIQYDRRRGTLVGALIVDDDTELYAI
Mvan_0007|M.vanbaalenii_PYR-1 SGGYAKRTAIEEYSAQGRGGKGILTIQYDRRRGTLVGALIVDDDTELYAI
TH_0756|M.thermoresistible__bu SGGYAKRTPIEEYPVQNRGGKGVLTIQYDRRRGNLVGALVVDDETELYAI
MSMEG_0006|M.smegmatis_MC2_155 SGGYAKRTSIDEYSVQGRGGKGILTIQYDRKRGSLVGALIVDDDTELYAI
:***:*** *:**. *.*****:**: ** :** ****::**:::****:
Mb0006|M.bovis_AF2122/97 TSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEESGDDNA
Rv0006|M.tuberculosis_H37Rv TSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEESGDDNA
MLBr_00006|M.leprae_Br4923 TSGGGVIRTTARQVRQAGRQTKGVRLMNLGEGDTLLAIARNAEESADGGV
MAV_0006|M.avium_104 TSGGGVIRTAAGQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEEAADEAV
MMAR_0006|M.marinum_M TSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEENADEAV
MUL_0006|M.ulcerans_Agy99 TSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLLAIARNAEENADEAV
MAB_0019|M.abscessus_ATCC_1997 TSGGGVIRTIAKQVRKAGRQTKGVRLMNLGEGDTLLAIAHNADEGDADPD
Mflv_0821|M.gilvum_PYR-GCK TSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLIAIARNAEAGDSTDE
Mvan_0007|M.vanbaalenii_PYR-1 TSGGGVIRTAARQVRKAGRQTKGVRLMNLGEGDTLIAIARNAEAGDSTDE
TH_0756|M.thermoresistible__bu TSSGGVIRTTARQIRKAGRQTKGVRLMNLAEGDTLVAIARNAEAGDSTDE
MSMEG_0006|M.smegmatis_MC2_155 TSTGGVIRTAARQVRKAGRQTKGVRLMNLAEGDTLIAIARNADEDEAAES
** ****** * *:*:*************.*****:***:**:
Mb0006|M.bovis_AF2122/97 VDANGADQTGN---
Rv0006|M.tuberculosis_H37Rv VDANGADQTGN---
MLBr_00006|M.leprae_Br4923 G-------------
MAV_0006|M.avium_104 EESDGAAGSDG---
MMAR_0006|M.marinum_M AEATDTDESSN---
MUL_0006|M.ulcerans_Agy99 AEATDTDESSN---
MAB_0019|M.abscessus_ATCC_1997 EDAAGTTAGE----
Mflv_0821|M.gilvum_PYR-GCK VE-TDPDATDEES-
Mvan_0007|M.vanbaalenii_PYR-1 VN-TDPDATDDEES
TH_0756|M.thermoresistible__bu VN-TDPDSV-----
MSMEG_0006|M.smegmatis_MC2_155 ISESDADTAESPEA