For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRPLTRRGFLAVTAGLALVAACSTDKPGEVADDGSVTVRHAFGDTTIPGPPQRVVSAGLTEQDDLLAVGV VPIAVTDWFGGEPFGVWPWAQRQLAGAQPAVLNLDNGIPVEEIAALKPDLIVATNAGLDADTYAKLSEIA PTVAQTGSEAFFEPWKDQATIIGQAVFKNAEMTELIKSVDDRFTTVKTDHPQFSGKKALLLGGTLYRGGV QATPPGWRTDFLTQMGLTVLQVPALIPRDEIASVLDGADVLIWTTESDQDRDALLADPIVAQLAATRRDR NIFTTKELAGAIAFASPLSYPVVADQLPPELARVLG*
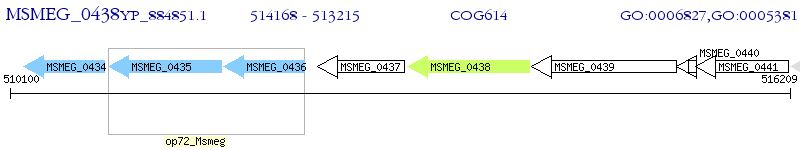
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0438 | - | - | 100% (317) | periplasmic binding protein |
| M. smegmatis MC2 155 | MSMEG_0020 | - | 4e-36 | 32.79% (308) | periplasmic binding protein |
| M. smegmatis MC2 155 | MSMEG_2319 | - | 1e-06 | 27.37% (285) | periplasmic binding protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0271c | - | 1e-106 | 61.42% (324) | periplasmic IRON-transport lipoprotein |
| M. gilvum PYR-GCK | Mflv_0375 | - | 1e-114 | 65.06% (312) | periplasmic binding protein |
| M. tuberculosis H37Rv | Rv0265c | - | 1e-106 | 61.42% (324) | periplasmic IRON-transport lipoprotein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4390 | - | 2e-86 | 50.30% (328) | ABC transporter periplasmic substrate-binding protein |
| M. marinum M | MMAR_0525 | - | 1e-107 | 60.92% (325) | periplasmic iron-transport lipoprotein |
| M. avium 104 | MAV_4895 | - | 1e-108 | 62.78% (317) | periplasmic binding protein |
| M. thermoresistible (build 8) | TH_1824 | fecB | 3e-08 | 29.51% (183) | PROBABLE FEIII-DICITRATE-BINDING PERIPLASMIC LIPOPROTEIN |
| M. ulcerans Agy99 | MUL_1188 | - | 1e-107 | 61.23% (325) | periplasmic iron-transport lipoprotein |
| M. vanbaalenii PYR-1 | Mvan_0308 | - | 1e-118 | 68.69% (313) | periplasmic binding protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0375|M.gilvum_PYR-GCK -----------------------MWSRRSFLGLTAGA---AATLTACGQD
Mvan_0308|M.vanbaalenii_PYR-1 --------------------MATLVPRRRFLTMSAGA---AALLTACGSE
MSMEG_0438|M.smegmatis_MC2_155 --------------------MPRPLTRRGFLAVTAG----LALVAACSTD
Mb0271c|M.bovis_AF2122/97 --------------------MRQGCSRRGFLQVAEAAAATG-LFAGCSSP
Rv0265c|M.tuberculosis_H37Rv --------------------MRQGCSRRGFLQVAEAAAATG-LFAGCSSP
MMAR_0525|M.marinum_M --------------------MQQGWSRRGFLQVAAATGLLASGVAACSSS
MUL_1188|M.ulcerans_Agy99 --------------------MQQGWSRRGFLQVAAATGLLASGVAACSSS
MAV_4895|M.avium_104 --------------------------------MAATAGAAG-LSAGCSAH
MAB_4390|M.abscessus_ATCC_1997 --------------------MPPGMITRRGLLAAGAALAASTALASCAKE
TH_1824|M.thermoresistible__bu VPLEPGPDRRRGGVTVGDVIVPGRRIRVIAGMVSLVTAVFLTMATGCGAP
: :.*.
Mflv_0375|M.gilvum_PYR-GCK K-------------------------------------PGT-VAGDGSVT
Mvan_0308|M.vanbaalenii_PYR-1 K-------------------------------------PGA-IASDGSVT
MSMEG_0438|M.smegmatis_MC2_155 K-------------------------------------PGE-VADDGSVT
Mb0271c|M.bovis_AF2122/97 K-------------------------------------PPPGTPGGAAVT
Rv0265c|M.tuberculosis_H37Rv K-------------------------------------PPPGTPGGAAVT
MMAR_0525|M.marinum_M K-------------------------------------PQAGTADTGAVT
MUL_1188|M.ulcerans_Agy99 K-------------------------------------PQAGTADTGAVT
MAV_4895|M.avium_104 Q-------------------------------------PPPGGAGPGSVT
MAB_4390|M.abscessus_ATCC_1997 S-------------------------------------APT-SERDGTAF
TH_1824|M.thermoresistible__bu QGAEPAPPRETVITSTTEIAGAGVLGNQRRPDESCAAEPAAPDPGPPVRD
. .
Mflv_0375|M.gilvum_PYR-GCK VAHVFGETKIPVPPTRVVSAGLTDADDLLALGVVPIAVTDWFGAEPFGVW
Mvan_0308|M.vanbaalenii_PYR-1 VRHAFGETKIPKPPTRVVSAGLTDADDLLALGVVPIAVTDWFGAEPFGVW
MSMEG_0438|M.smegmatis_MC2_155 VRHAFGDTTIPGPPQRVVSAGLTEQDDLLAVGVVPIAVTDWFGGEPFGVW
Mb0271c|M.bovis_AF2122/97 ITHLFGQTVIKEPPKRVVSAGYTEQDDLLAVDVVPIAVTDWFGDQPFAVW
Rv0265c|M.tuberculosis_H37Rv ITHLFGQTVIKEPPKRVVSAGYTEQDDLLAVDVVPIAVTDWFGDQPFAVW
MMAR_0525|M.marinum_M ITHLFGQTVIKEPPKRVVSAGFTEQDDLLALGVVPIAVTRWFGDQPFAVW
MUL_1188|M.ulcerans_Agy99 ITHLFGQTVIKEPPKRVVSAGFTEQDDLLALGVVPIAVTRWFGDQPFAVW
MAV_4895|M.avium_104 VTHLFGQTVVKEPPKRVVSAGYTEQDDLLAVGVVPIAVTNWFGDQPFAVW
MAB_4390|M.abscessus_ATCC_1997 IKHAFGTTEVKAPPKRIISAGYTGHDYLLALGIVPVAVTEWYGGFPFATW
TH_1824|M.thermoresistible__bu VAHAAGSTAVPADPQRIVVLSGDQLDALCALGLQSRIVAAAVP-EDTGVQ
: * * * : * *:: . * * *:.: . *: ..
Mflv_0375|M.gilvum_PYR-GCK PWARPQLGAAQPVVLSLADGVQVEQIASLNPDLIIATDAGLDQDTYTRLS
Mvan_0308|M.vanbaalenii_PYR-1 PWAQPRLGGAQPVVLSLADGIQVEQIAGLTPDLIVATDAGLDQDTYTRLS
MSMEG_0438|M.smegmatis_MC2_155 PWAQRQLAGAQPAVLNLDNGIPVEEIAALKPDLIVATNAGLDADTYAKLS
Mb0271c|M.bovis_AF2122/97 PWAAPKLGGARPAVLNLDNGIQIDRIAALKPDLIVAINAGVDADTYQQLS
Rv0265c|M.tuberculosis_H37Rv PWAAPKLGGARPAVLNLDNGIQIDRIAALKPDLIVAINAGVDADTYQQLS
MMAR_0525|M.marinum_M PWAQPKLGAAQPVVLSLDDGIQVDQIANLKPDLIVAINAGVDAETYQKLS
MUL_1188|M.ulcerans_Agy99 PWAQPKLGAAQPVVLSLDDGIQVDQIANLKPDLIVAINAGVDAETYQKLS
MAV_4895|M.avium_104 PWAADKLGGAQPTVLNLDNGIPVDQIAGLKPDLIVAINAGLDADTYQKLS
MAB_4390|M.abscessus_ATCC_1997 PWATERLGGATPEVLTLTDGLMIDKIASLKPDLIVAADAGLDQDTYTKLS
TH_1824|M.thermoresistible__bu PAYLGTVIHELPAAG-TRTEPDLDAIRAARPDLILGSQA-LTPADYPRLA
* : * . :: * ****:. :* : * :*:
Mflv_0375|M.gilvum_PYR-GCK QIAPTVAQSGRAAFFEPWRDQATAIGQAVFRHDDMQKLIA--DVDAQFSA
Mvan_0308|M.vanbaalenii_PYR-1 EIAPTIAQSGSAAFFEPWRDQAGAIGQAVLRHDDMQKLIT--DVDAKFTG
MSMEG_0438|M.smegmatis_MC2_155 EIAPTVAQTGSEAFFEPWKDQATIIGQAVFKNAEMTELIK--SVDDRFTT
Mb0271c|M.bovis_AF2122/97 AIAPTVAQSGGDAFFEPWKDQARSIGQAVFAADRMRSLIE--AVDQKFAA
Rv0265c|M.tuberculosis_H37Rv AIAPTVAQSGGDAFFEPWKDQARSIGQAVFAADRMRSLIE--AVDQKFAA
MMAR_0525|M.marinum_M AIAPTIPQSGGDAFFEPWKLQASTVGEAVFQSAQMKSLID--AVGQKFAA
MUL_1188|M.ulcerans_Agy99 AIAPTIPQSGGDAFFEPWKLQASTVGEAVFQRAQMKSLID--AVDQKFAA
MAV_4895|M.avium_104 AIAPTVAQSDGDAFFEPWKEQAAAVGQAVFAAEKMKSLVA--GVDQKFTD
MAB_4390|M.abscessus_ATCC_1997 GIAPTIAQAGKYAFFEPWKDQAKAIGQAVFKPAEMDAVVK--AVDNKFSS
TH_1824|M.thermoresistible__bu AIAPTVFTG---APGPDWQGNLRTVGAATGRSAAADALLDGFAEDARRTG
****: * *: : :* *. :: . : :
Mflv_0375|M.gilvum_PYR-GCK AKDAHPEFSGKTALLLAGSVTGDGVRVTGPAWRREF-LTAMGLTVLD---
Mvan_0308|M.vanbaalenii_PYR-1 VKDSHPQFAGKKALLLAGTLFRDSVQVSGPSWRHEF-LTQMGLTVLE---
MSMEG_0438|M.smegmatis_MC2_155 VKTDHPQFSGKKALLLGGTLYRGGVQATPPGWRTDF-LTQMGLTVLQ---
Mb0271c|M.bovis_AF2122/97 VAQRHPRWRGKKALLLQGRLWQGNVVATLAGWRTDF-LNDMGLVIADSIK
Rv0265c|M.tuberculosis_H37Rv VAQRHPRWRGKKALLLQGRLWQGNVVATLAGWRTDF-LNDMGLVIADSIK
MMAR_0525|M.marinum_M VAQQHPQWMGKKALLLQGRLWQGNVIATMAGWRTDF-LNEMGLVISEGIK
MUL_1188|M.ulcerans_Agy99 AAQQHPQWMGKKALLLQGRLWQGNVIATMAGWRTDF-LNEMGLVVSEGIK
MAV_4895|M.avium_104 IGKKNPQWTGKKALLMHGALWQGTVVATMAGWRTDF-LNQMGLVIADSIK
MAB_4390|M.abscessus_ATCC_1997 AASAHTGFKDKKAIYLQGELLDGQAIAFRSGPRTGF-LSGLGLITPPDLE
TH_1824|M.thermoresistible__bu AQNDAAHFQASVVQLTDTTVRVFGADTFPGSVLAAVGLDRPAAQRFTDRP
. : . . : . . . . * .
Mflv_0375|M.gilvum_PYR-GCK ----TDD--EIPRDRMPDVLDEADVLIWTTESDEEQASLLADPAIADLRA
Mvan_0308|M.vanbaalenii_PYR-1 ----TDDA-LIPRDRIAEVLGAADVLIWTTESDQEQAALLADPAIAGIPA
MSMEG_0438|M.smegmatis_MC2_155 ----VPA--LIPRDEIASVLDGADVLIWTTESDQDRDALLADPIVAQLAA
Mb0271c|M.bovis_AF2122/97 PFA-VDQRGVIPRDHIKAVLDAADVLIWMTESPEDEKALLADPEIAASQA
Rv0265c|M.tuberculosis_H37Rv PFA-VDQRGVIPRDHIKAVLDAADVLIWMTESPEDEKALLADPEIAASQA
MMAR_0525|M.marinum_M PFA-VDQRGVIPRDHIKEVLDSADVLIWLTESPEDEQALLADPEIAASAA
MUL_1188|M.ulcerans_Agy99 PFA-VDQRGVIPRDHIKEVLDSADVLIWLTESPEDEQALLADPEIAASAA
MAV_4895|M.avium_104 PFG-TDQRAVIPRDHIKSVLESADVVIWTTQNPDDQKALLADPEVAGSLT
MAB_4390|M.abscessus_ATCC_1997 TYAKGDQLAYLAADRLSTVLAEADVLLWGTESDEQIAALKADPAITTLGP
TH_1824|M.thermoresistible__bu YIDIGATDAELADADFGIAEGDIVYVSFASPAARDRARTVFTSDAWRRLS
:. : . : : : : . .
Mflv_0375|M.gilvum_PYR-GCK TTRSSHVFTGKELAGAIAYASTLSYPVLADRLPPMLADALT-
Mvan_0308|M.vanbaalenii_PYR-1 RR---HVFTGKELAGAIAYASTLSYPVVADRLPPMLAAAVG-
MSMEG_0438|M.smegmatis_MC2_155 TRRDRNIFTTKELAGAIAFASPLSYPVVADQLPPELARVLG-
Mb0271c|M.bovis_AF2122/97 TAQRRHIFTSKEQAGAIAFSSVLSYPVVAEQLPPQISQILGA
Rv0265c|M.tuberculosis_H37Rv TAQRRHIFTSKEQAGAIAFSSVLSYPVVAEQLPPQISQILGA
MMAR_0525|M.marinum_M TEQKRHVFTTGDQAGAIAFASPLSYPVVADELPALIAKVLG-
MUL_1188|M.ulcerans_Agy99 TEQKRHVFTTGDQAGAIAFASPLSYPVVADELPALIAKVLG-
MAV_4895|M.avium_104 TAQNRHIFTTKDQAGAIAFASPLSYPVIADQLPPQLTKILG-
MAB_4390|M.abscessus_ATCC_1997 SLLDRSIFTSKELGGAINFSSPLSLTYVVDNLVPQLARILG-
TH_1824|M.thermoresistible__bu ATKDNRVFTVNNEVWQTGQN-IVAARGILDDLQWVNAPIN--
:** : :: : : * :