For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FYGCRVSDTVIRRAEPADVPGIERLVRDAFALYVPRIGREPAPMSADYAAALENSRVWVLEADGRLVGAL VNEVHDDHLLLDTVAVAPGTQGRGYGARLLARAEQDARELGLPEVRLYTNEKMTENQTFYPRHGYVETAR GEQDGYSRVFYAKRITR*
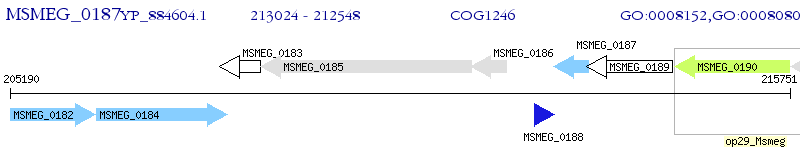
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_0187 | - | - | 100% (158) | acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_2691 | - | 1e-09 | 36.72% (128) | N-acetylglutamate synthase |
| M. smegmatis MC2 155 | MSMEG_2405 | - | 9e-07 | 32.43% (111) | MarR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2768 | - | 1e-06 | 35.61% (132) | N-acetylglutamate synthase |
| M. gilvum PYR-GCK | Mflv_3990 | - | 1e-07 | 34.11% (129) | N-acetylglutamate synthase |
| M. tuberculosis H37Rv | Rv2747 | - | 1e-06 | 35.61% (132) | N-acetylglutamate synthase |
| M. leprae Br4923 | MLBr_00978 | - | 5e-06 | 32.58% (132) | N-acetylglutamate synthase |
| M. abscessus ATCC 19977 | MAB_1138c | - | 5e-08 | 34.15% (82) | MarR family transcriptional regulator |
| M. marinum M | MMAR_1968 | - | 2e-05 | 33.33% (132) | acetyltransferase |
| M. avium 104 | MAV_3562 | - | 2e-06 | 31.31% (99) | acetyltransferase, gnat family protein |
| M. thermoresistible (build 8) | TH_0578 | - | 2e-08 | 35.16% (128) | POSSIBLE TRANSFERASE |
| M. ulcerans Agy99 | MUL_3389 | - | 1e-05 | 33.33% (132) | N-acetylglutamate synthase |
| M. vanbaalenii PYR-1 | Mvan_2394 | - | 9e-07 | 32.56% (129) | N-acetylglutamate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
TH_0578|M.thermoresistible__bu -----------------VRR----------ARTSDVPAIKALVD------
Mvan_2394|M.vanbaalenii_PYR-1 ----MSADPDAATGQLLVRR----------ARTSDVPAIKALVD------
Mflv_3990|M.gilvum_PYR-GCK ----------------MVRR----------ARTSDVPHIKALVD------
Mb2768|M.bovis_AF2122/97 ------MTERPRDCRPVVRR----------ARTSDVPAIKQLVD------
Rv2747|M.tuberculosis_H37Rv ------MTERPRDCRPVVRR----------ARTSDVPAIKQLVD------
MMAR_1968|M.marinum_M ------MTESQQDFRPVVRR----------ARTSDVPAIKKLVD------
MUL_3389|M.ulcerans_Agy99 ------MTESQQDFRPVVRR----------ARTSDVPAIKKLVD------
MLBr_00978|M.leprae_Br4923 MSILLAVTKNSQNYLPVVRR----------ARTSDIPAIKQLVD------
MSMEG_0187|M.smegmatis_MC2_155 ------MFYGCRVSDTVIRR----------AEPADVPGIERLVRDA----
MAB_1138c|M.abscessus_ATCC_199 --MTDALVDGLRSFNRTVTQRIGVLESEYLARDRPLGQSRVLWEIGDSGC
MAV_3562|M.avium_104 ---MESLTPAPATQAVRIVT----------TESVDVQQLADLAAR-----
: :. : *
TH_0578|M.thermoresistible__bu ---IYAGRILLEKN-LVTLYES-----------VQEFWVAE---RDGELV
Mvan_2394|M.vanbaalenii_PYR-1 ---IYAGKILLEKN-LVTLYES-----------VQEFWVAE---IDGELI
Mflv_3990|M.gilvum_PYR-GCK ---IYAGRILLEKN-LVTLYES-----------VQEFWVAE---VDGEVV
Mb2768|M.bovis_AF2122/97 ---TYAGKILLEKN-LVTLYEA-----------VQEFWVAEHPDLYGKVV
Rv2747|M.tuberculosis_H37Rv ---TYAGKILLEKN-LVTLYEA-----------VQEFWVAEHPDLYGKVV
MMAR_1968|M.marinum_M ---TYAGKILLEKN-LVTLYEA-----------VQEFWVAEHPEIQGNVI
MUL_3389|M.ulcerans_Agy99 ---TYAGKILLEKN-LVTLYEA-----------VQEFWVAEHPEIQGNVI
MLBr_00978|M.leprae_Br4923 ---TYAGKILLEKN-LVTLYEA-----------VQEFWVAEHPDIPGKVV
MSMEG_0187|M.smegmatis_MC2_155 -FALYVPRIGREPAPMSADYAA-----------ALENSRVWVLEADGRLV
MAB_1138c|M.abscessus_ATCC_199 DIRELRARLGLDSGYLSRLLRA-----------LEGAGLIEVTAADGDTR
MAV_3562|M.avium_104 -TFPLACPTSAAPEDIAAFIDANLSAECFARYLADPHRAVLAAELDGRIL
: : *
TH_0578|M.thermoresistible__bu GCGALHVMWS-DLGEVRTVAVHPKVRGQGVGHMIVERLLDVARELELERI
Mvan_2394|M.vanbaalenii_PYR-1 GCGALHVMWS-DLGEVRTVAVHPKALGRGVGHAIVERLLSVAADLHLRRI
Mflv_3990|M.gilvum_PYR-GCK GCGALHVLWS-DLGEVRTVAVDPKVRGRGVGHAIVERLLTVAADLRLQRI
Mb2768|M.bovis_AF2122/97 GCGALHVLWS-DLGEIRTVAVDPAMTGHGIGHAIVDRLLQVARDLQLQRV
Rv2747|M.tuberculosis_H37Rv GCGALHVLWS-DLGEIRTVAVDPAMTGHGIGHAIVDRLLQVARDLQLQRV
MMAR_1968|M.marinum_M GCGALHVLWS-DLGEVRTVAVDPAMTGHGIGHAIVDRLLEVARELQLERL
MUL_3389|M.ulcerans_Agy99 GCGALHVLWS-DLGEVRTVAVDPAMTGHGIGHAIVDRLLEVARELQLERL
MLBr_00978|M.leprae_Br4923 GCGALHVLWS-DLGEIRTVAVDPAMTGHGVGGTIVNRLLEVARKLELERL
MSMEG_0187|M.smegmatis_MC2_155 GA-LVNEVHD-DHLLLDTVAVAPGTQGRGYGARLLARAEQDARELGLP--
MAB_1138c|M.abscessus_ATCC_199 VR-TARLTAA-GRAELAELDRRSDDLAHSILEPLSVNQRQRLADAMAEVE
MAV_3562|M.avium_104 GYAMLIHDLAGDAAELSKIYVAPEQHGAGTAAALMRLALDTAGRWGATRV
. : : . . . :
TH_0578|M.thermoresistible__bu FVLTFEVEFFARHGFREIEGTPVTAEVYEEMCRSYDTGVAEFLDLSYVKP
Mvan_2394|M.vanbaalenii_PYR-1 FVLTFEVEFFGRHGFQEIEGTPVTSEVYEEMCRSYDTGVAEFLDLSYVKP
Mflv_3990|M.gilvum_PYR-GCK FVLTFEVDFFARHGFQEIEGTPVTAEVYEEMCRSYDTGVAEFLDLSYVKP
Mb2768|M.bovis_AF2122/97 FVLTFETEFFARHGFTEIEGTPVTAEVFDEMCRSYDIGVAEFLDLSYVKP
Rv2747|M.tuberculosis_H37Rv FVLTFETEFFARHGFTEIEGTPVTAEVFDEMCRSYDIGVAEFLDLSYVKP
MMAR_1968|M.marinum_M FVLTFETEFFTAHGFTEIEGTPVTAEVFEEMCRSYDIGVAEFLDLSYIKP
MUL_3389|M.ulcerans_Agy99 FVLTFETEFFTAHGFTEIEGTPVTAEVFEEMCRSYDIGVAEFLDLSYIKP
MLBr_00978|M.leprae_Br4923 FVLTFETEFFSQHGFTEIEGTPVTAEVFEEMCRSYDIGVAEFLDLSYVKP
MSMEG_0187|M.smegmatis_MC2_155 -----EVRLYTNEKMTENQ-TFYPRHGYVETARGEQDGYSRVFYAKRITR
MAB_1138c|M.abscessus_ATCC_199 RLFVASQVQIAVMDTRTPEARYCIRAYFEEISARFEDGFDPALTLPAGDA
MAV_3562|M.avium_104 WLGVNQKNQRAQR-FYAKHGFTVSGTRTFQLGAGREDDYLMSRTLR----
. . : : .
TH_0578|M.thermoresistible__bu NILGNTRMLLNL--------------------------------------
Mvan_2394|M.vanbaalenii_PYR-1 NILGNTRMLLTL--------------------------------------
Mflv_3990|M.gilvum_PYR-GCK NILGNTRMLLLL--------------------------------------
Mb2768|M.bovis_AF2122/97 NILGNSRMLLVL--------------------------------------
Rv2747|M.tuberculosis_H37Rv NILGNSRMLLVL--------------------------------------
MMAR_1968|M.marinum_M NILGNTRMLLVL--------------------------------------
MUL_3389|M.ulcerans_Agy99 NILGNTRMLLVL--------------------------------------
MLBr_00978|M.leprae_Br4923 NTLGNSRMLLVL--------------------------------------
MSMEG_0187|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1138c|M.abscessus_ATCC_199 ELCAPAGLFLVAALHEEPVGCAGLKLPPHTDTAEIKRVWIAPHVRGLGLG
MAV_3562|M.avium_104 --------------------------------------------------
TH_0578|M.thermoresistible__bu --------------------------------------------------
Mvan_2394|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_3990|M.gilvum_PYR-GCK --------------------------------------------------
Mb2768|M.bovis_AF2122/97 --------------------------------------------------
Rv2747|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_1968|M.marinum_M --------------------------------------------------
MUL_3389|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00978|M.leprae_Br4923 --------------------------------------------------
MSMEG_0187|M.smegmatis_MC2_155 --------------------------------------------------
MAB_1138c|M.abscessus_ATCC_199 KRILAELERLAVDHGAHTIRLDTNRALTEAIAMYRSAGYDEVAAFNQEKY
MAV_3562|M.avium_104 --------------------------------------------------
TH_0578|M.thermoresistible__bu ----------
Mvan_2394|M.vanbaalenii_PYR-1 ----------
Mflv_3990|M.gilvum_PYR-GCK ----------
Mb2768|M.bovis_AF2122/97 ----------
Rv2747|M.tuberculosis_H37Rv ----------
MMAR_1968|M.marinum_M ----------
MUL_3389|M.ulcerans_Agy99 ----------
MLBr_00978|M.leprae_Br4923 ----------
MSMEG_0187|M.smegmatis_MC2_155 ----------
MAB_1138c|M.abscessus_ATCC_199 AHFWFQKQLS
MAV_3562|M.avium_104 ----------