For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MADPQDRPDGEPDATPGQPAKKPPADKAAKTPAKKTPAKKTPAKKAPAQKAPAKKAPAKKAPAKKAPATK PPAQSPEPAPQPTALANVSQQQPSANGQLAEAAKDAAAQAKSTVDRANNPLPRVADEQPSHSTVPLVVAI ALSLLAMLLIRQLRRR
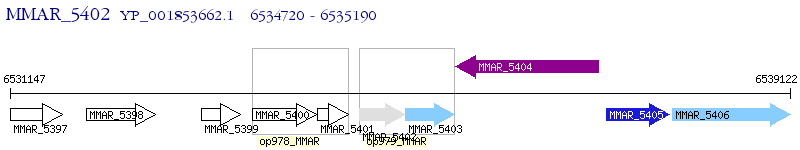
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5402 | hns | - | 100% (156) | histone-like protein Hns |
| M. marinum M | MMAR_3834 | plsC | 1e-17 | 50.43% (115) | bifunctional transmembrane phospholipid biosynthesis enzyme |
| M. marinum M | MMAR_1728 | hupB | 4e-10 | 45.10% (102) | DNA-binding protein HU, HupB |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3882 | hns | 3e-37 | 58.60% (157) | histone-like protein HNS |
| M. gilvum PYR-GCK | Mflv_1125 | - | 2e-19 | 40.62% (160) | hypothetical protein Mflv_1125 |
| M. tuberculosis H37Rv | Rv3852 | hns | 3e-37 | 58.60% (157) | histone-like protein HNS |
| M. leprae Br4923 | MLBr_00067 | hns | 2e-19 | 40.51% (158) | histone-like protein |
| M. abscessus ATCC 19977 | MAB_0108c | - | 7e-20 | 38.46% (195) | hypothetical protein MAB_0108c |
| M. avium 104 | MAV_0176 | - | 2e-35 | 54.78% (157) | hypothetical protein MAV_0176 |
| M. smegmatis MC2 155 | MSMEG_6438 | - | 5e-29 | 47.53% (162) | hypothetical protein MSMEG_6438 |
| M. thermoresistible (build 8) | TH_2451 | - | 2e-23 | 43.98% (166) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_5024 | hns | 5e-84 | 98.11% (159) | histone-like protein Hns |
| M. vanbaalenii PYR-1 | Mvan_5681 | - | 1e-23 | 43.48% (161) | hypothetical protein Mvan_5681 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5402|M.marinum_M MADP---QDRPDGEPDATPG------------------QPAKKP----PA
MUL_5024|M.ulcerans_Agy99 MADP---QDRPDGEPDATPG------------------QPAKKP----PA
Mb3882|M.bovis_AF2122/97 MPDP---QDRPDSEPSDAST------------------PPAKKL----PA
Rv3852|M.tuberculosis_H37Rv MPDP---QDRPDSEPSDAST------------------PPAKKL----PA
MLBr_00067|M.leprae_Br4923 MADQ---QNPPNSEPDGTST------------------LPAKKV----PA
MAV_0176|M.avium_104 MADP---QDPTNSAADGAGD------------------PPEKKP----PA
TH_2451|M.thermoresistible__bu MADP---QDQPAGTGDSVSS------------------GPDPAAGSPEPA
Mflv_1125|M.gilvum_PYR-GCK MSDQ---PDGPDESPATPTP-----PATPP--------PPVKKA----PA
Mvan_5681|M.vanbaalenii_PYR-1 MADE---PDRPDESPDTATP-----ATPPPV-------PPVKKA----AA
MSMEG_6438|M.smegmatis_MC2_155 MAEQ---QDRPTENPQTPSG-----DSAAGASGSGDNSAPAKKA----PA
MAB_0108c|M.abscessus_ATCC_199 MADQDNSANEPNQTPEKAAGGESPKPSAPARPAKAAKRPAAKRA----PA
*.: : . . .*
MMAR_5402|M.marinum_M DKAAKTPAKKTPAKKTPA------------------KKAPAQKAP---AK
MUL_5024|M.ulcerans_Agy99 DKAAKTPAKKTPAKKTPA------------------KKAPAQKAP---AK
Mb3882|M.bovis_AF2122/97 KKAAK--------------------------------KAPARKTP---AK
Rv3852|M.tuberculosis_H37Rv KKAAK--------------------------------KAPARKTP---AK
MLBr_00067|M.leprae_Br4923 K-----------------------------------------------AA
MAV_0176|M.avium_104 KAAKKT------------------------------AKAPAKKAP---AK
TH_2451|M.thermoresistible__bu EKAGAPAEKKTERAQKAT------------------KKTPAQKTP---AK
Mflv_1125|M.gilvum_PYR-GCK KKAPAKKAPAKKAT-------------------------PAKAAP---VK
Mvan_5681|M.vanbaalenii_PYR-1 RKA----APSKKAT-------------------------PAKKVP---AK
MSMEG_6438|M.smegmatis_MC2_155 KKSPAEKAPAKKA--------------------------PAKKAPAKAAK
MAB_0108c|M.abscessus_ATCC_199 KRPAGSSAPPKSGPKAAPAPSADAAPKPPAAAKPAEAPKPAAPAPAPAAE
.
MMAR_5402|M.marinum_M KAPAKKAPAKKAPATKPPAQSPE---PAPQPTALANVSQQQPSANGQLAE
MUL_5024|M.ulcerans_Agy99 KAPAKKAPAKKAPATKPPAQSPEPAEPAPQPTALANVSQQQPSANGQLAE
Mb3882|M.bovis_AF2122/97 KAPAKKTPAKGAKSAPPKP--------AEAPVSL----QQRIETNGQLAA
Rv3852|M.tuberculosis_H37Rv KAPAKKTPAKGAKSAPPKP--------AEAPVSL----QQRIETNGQLAA
MLBr_00067|M.leprae_Br4923 KAPAKKAPPKSPSFASPQA--------ANQSLGL----QQRIETNGQLD-
MAV_0176|M.avium_104 KAPAKKAPAKKAPAKKAPA--------KNTPTGG----GQRADTNGDLTA
TH_2451|M.thermoresistible__bu KA-AQKTPAKKATKKAPAQKAP-----ARKPAEPERRQTAGAAPPADLTS
Mflv_1125|M.gilvum_PYR-GCK KAPAKKAPVKKAAAKKAPP----------------------PPADA-VTS
Mvan_5681|M.vanbaalenii_PYR-1 KVPAKKAPVKKAAAKKAAPT-------KAAPTKE------APPVQASLAA
MSMEG_6438|M.smegmatis_MC2_155 KAPAKKAPAKKAPAKKAEPA-------APAPVEKP----TAAPVDE-IAA
MAB_0108c|M.abscessus_ATCC_199 PAPAPKPAAQPVSEAPAAPK-------PVAPASKPE-AAKPEPAQPDLAA
. * *.. : . :
MMAR_5402|M.marinum_M AAKDAAAQAKSTVDRANNPLPR--VADE-QPSHSTVPLVVAIALSLLAML
MUL_5024|M.ulcerans_Agy99 AAKDAAAQAKSTVDRANNPLPR--VADE-QPSHSTVPLVVAIALSLLAML
Mb3882|M.bovis_AF2122/97 AAKDAAAQAKSTVEGANDALAR--NASVPAPSHSPVPLIVAVTLSLLALL
Rv3852|M.tuberculosis_H37Rv AAKDAAAQAKSTVEGANDALAR--NASVPAPSHSPVPLIVAVTLSLLALL
MLBr_00067|M.leprae_Br4923 VAKDVAEQAQSAVEGASNPVPN--GAEALEASNSPVALVIALAIGLLALL
MAV_0176|M.avium_104 AAKDAAAQAKSTVEAADNPVAP--AVVGPAAGQSPVPLIVAVAVSLLAIL
TH_2451|M.thermoresistible__bu AAKEVAAHAKSTVASAPNPVAT--PAPISRPDDSRLPFMIAVASGALILL
Mflv_1125|M.gilvum_PYR-GCK AAKEAAAQAKSTVATASTAVTSPPPAPPGPASSSRLPLAAAAVAGVLALV
Mvan_5681|M.vanbaalenii_PYR-1 AAEETAAQAKSTVATASTAVSAPPPAPPAGLDSSRLPIVAALVAGVLALV
MSMEG_6438|M.smegmatis_MC2_155 AAEEVAAQAKSTVEAAPNPVSGPVPVPVEEHHSTRFPVGPTIAAAVMIVL
MAB_0108c|M.abscessus_ATCC_199 AARETAAQAKSTVDSAPPPIPG--PAPEQGRQSPNLKIAVGAVAAILALL
.*.:.* :*:*:* * .:. . . . . . . : ::
MMAR_5402|M.marinum_M LIRQLRRR-------
MUL_5024|M.ulcerans_Agy99 LIRQLRRR-------
Mb3882|M.bovis_AF2122/97 LIRQLRRR-------
Rv3852|M.tuberculosis_H37Rv LIRQLRRR-------
MLBr_00067|M.leprae_Br4923 LIHQLRRR-------
MAV_0176|M.avium_104 LIRQLRRR-------
TH_2451|M.thermoresistible__bu IVLLVNRRGK-----
Mflv_1125|M.gilvum_PYR-GCK VALLVARRNSDD---
Mvan_5681|M.vanbaalenii_PYR-1 VILLAARRNNDR---
MSMEG_6438|M.smegmatis_MC2_155 LVLAARRRGAQD---
MAB_0108c|M.abscessus_ATCC_199 LLRRRRRREDSVESE
: **