For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SDQPDGPDESPATPTPPATPPPPVKKAPAKKAPAKKAPAKKATPAKAAPVKKAPAKKAPVKKAAAKKAPP PPADAVTSAAKEAAAQAKSTVATASTAVTSPPPAPPGPASSSRLPLAAAAVAGVLALVVALLVARRNSDD *
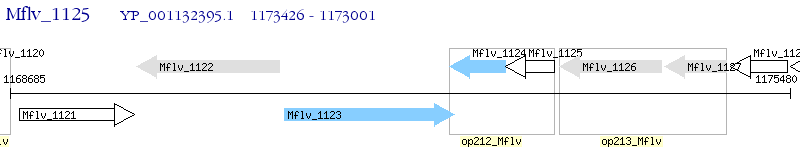
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1125 | - | - | 100% (141) | hypothetical protein Mflv_1125 |
| M. gilvum PYR-GCK | Mflv_4225 | - | 1e-14 | 47.93% (121) | histone family protein DNA-binding protein |
| M. gilvum PYR-GCK | Mflv_0100 | - | 1e-11 | 62.71% (59) | hypothetical protein Mflv_0100 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3882 | hns | 2e-18 | 42.11% (152) | histone-like protein HNS |
| M. tuberculosis H37Rv | Rv3852 | hns | 2e-18 | 42.11% (152) | histone-like protein HNS |
| M. leprae Br4923 | MLBr_00067 | hns | 2e-08 | 34.53% (139) | histone-like protein |
| M. abscessus ATCC 19977 | MAB_3292c | - | 3e-17 | 49.56% (113) | DNA-binding protein HU |
| M. marinum M | MMAR_3834 | plsC | 3e-20 | 57.30% (89) | bifunctional transmembrane phospholipid biosynthesis enzyme |
| M. avium 104 | MAV_0176 | - | 5e-21 | 48.53% (136) | hypothetical protein MAV_0176 |
| M. smegmatis MC2 155 | MSMEG_6438 | - | 5e-29 | 45.40% (163) | hypothetical protein MSMEG_6438 |
| M. thermoresistible (build 8) | TH_2451 | - | 2e-23 | 47.13% (157) | PUTATIVE - |
| M. ulcerans Agy99 | MUL_5024 | hns | 3e-19 | 39.88% (163) | histone-like protein Hns |
| M. vanbaalenii PYR-1 | Mvan_5681 | - | 3e-46 | 66.89% (151) | hypothetical protein Mvan_5681 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3834|M.marinum_M MSAAEEPGDGREQDKAAGDLRLPGSVAEIMASPAGPKVGAFFDLDGTLVA
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3834|M.marinum_M GFTAVILTQERLRRRDIGVGELLSMVQAGLNHTLGRIEFEQLINTASSAL
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3834|M.marinum_M RGRQLVDLEEIGERLFAQRIESRIYPEMRELVRAHVARGHTVVLSSSALT
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3834|M.marinum_M IQVNPVARFLGIVNMLTNKFETNEDGLLTGGVVKPILWGPGKAAAVQRFA
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3834|M.marinum_M AEHDIDLKDSYFYADGDEDVALMYLVGNPRPTNPEGKMAAVAKRRGWPIL
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 --------------------------------------------------
MMAR_3834|M.marinum_M KFNSRGGVGLRRQIRTLAGFSTMFPVAAGAVGIGLLTGNRRRGVNFFTSN
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 --------------------------------------------MNKAEL
MMAR_3834|M.marinum_M FSQLLLATSGVHLNVVGKENLTAQRPAVFIFNHRNQVDPVIAGALVRDNW
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3882|M.bovis_AF2122/97 --------------------------------------------------
Rv3852|M.tuberculosis_H37Rv --------------------------------------------------
MLBr_00067|M.leprae_Br4923 --------------------------------------------------
MAV_0176|M.avium_104 --------------------------------------------------
MUL_5024|M.ulcerans_Agy99 --------------------------------------------------
TH_2451|M.thermoresistible__bu --------------------------------------------------
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------------
MAB_3292c|M.abscessus_ATCC_199 IDVLTQKLGSD---------------RRQATAAVEHVVDTIVRTVHKGES
MMAR_3834|M.marinum_M VGVGKKELQNDPIMGTLGKVLDGVFIDRDDPASAVETLHTVEERAKNGLS
Mflv_1125|M.gilvum_PYR-GCK --------------------------------------------MSDQPD
Mvan_5681|M.vanbaalenii_PYR-1 --------------------------------------------MADEPD
Mb3882|M.bovis_AF2122/97 --------------------------------------------MPDPQD
Rv3852|M.tuberculosis_H37Rv --------------------------------------------MPDPQD
MLBr_00067|M.leprae_Br4923 --------------------------------------------MADQQN
MAV_0176|M.avium_104 --------------------------------------------MADPQD
MUL_5024|M.ulcerans_Agy99 --------------------------------------------MADPQD
TH_2451|M.thermoresistible__bu --------------------------------------------MADPQD
MSMEG_6438|M.smegmatis_MC2_155 --------------------------------------------MAEQQD
MAB_3292c|M.abscessus_ATCC_199 VTITGFGVFEQRRRAARVARN------------------------PRTGE
MMAR_3834|M.marinum_M IVIAPEGTRLDTTEVGPFKKGPFRIAMAAGIPIVPIVIRNAEIVASRNST
.
Mflv_1125|M.gilvum_PYR-GCK GPDESPATPTPPATPP--------PPVKKAPAKKAP--------AKKAPA
Mvan_5681|M.vanbaalenii_PYR-1 RPDESPDTATPATPPPV-------PPVKKAAARKA------------APS
Mb3882|M.bovis_AF2122/97 RPDSEPSDASTPPAKKL--------PAKKAAK--------------KAPA
Rv3852|M.tuberculosis_H37Rv RPDSEPSDASTPPAKKL--------PAKKAAK--------------KAPA
MLBr_00067|M.leprae_Br4923 PPNSEPDGTSTLPAKKV--------PAK----------------------
MAV_0176|M.avium_104 PTNSAADGAGDPPEKKP--------PAKAAKKT------------AKAPA
MUL_5024|M.ulcerans_Agy99 RPDGEPDATPGQPAKKP--------PADKAAKTPAKKTPAKKTPAKKAPA
TH_2451|M.thermoresistible__bu QPAGTGDSVSSGPDPAAG----SPEPAEKAGAPAEKKTERAQKATKKTPA
MSMEG_6438|M.smegmatis_MC2_155 RPTENPQTPSGDSAAGASGSGDNSAPAKKAPAK-------------KSPA
MAB_3292c|M.abscessus_ATCC_199 TVKVKPTSVPTFRPG-------AQFKAVVSGAQKLPADG---------PA
MMAR_3834|M.marinum_M TINPGTVDIAVFPPIPVDDWTLDTLPDRIAEVRQLYLDTLKDWPVDELPE
Mflv_1125|M.gilvum_PYR-GCK KKATPAKAAPVKKAPA---KKAPVKKAAA--------KKAPP--------
Mvan_5681|M.vanbaalenii_PYR-1 KKATPAKKVPAKKVPA---KKAPVKKAAA--------KKAAPTKAAPTKE
Mb3882|M.bovis_AF2122/97 RKT-PAKKAPAKKTPA---KGAKSAPPKP--------AEAPVSL----QQ
Rv3852|M.tuberculosis_H37Rv RKT-PAKKAPAKKTPA---KGAKSAPPKP--------AEAPVSL----QQ
MLBr_00067|M.leprae_Br4923 -----AAKAPAKKAPP---KSPSFASPQA--------ANQSLGL----QQ
MAV_0176|M.avium_104 KKA-PAKKAPAKKAPA---KKAPAKKAPA--------KNTPTGG----GQ
MUL_5024|M.ulcerans_Agy99 QKA-PAKKAPAKKAPA---KKAPATKPPAQSPEPAEPAPQPTALANVSQQ
TH_2451|M.thermoresistible__bu QKT-PAKKA-AQKTPA---KKATKKAPAQKAP-----ARKPAEPERRQTA
MSMEG_6438|M.smegmatis_MC2_155 EKA-PAKKAPAKKAPAKAAKKAPAKKAPAKKAP--AKKAEPAAPAPVEKP
MAB_3292c|M.abscessus_ATCC_199 VKR-GSTAAPAKRAAAK--KAAPAKKAPA--------KKAAPAKKAPVKK
MMAR_3834|M.marinum_M VNL-YAEEKAAKKAKAQ--LAKASAKETP--------AKKAPAKKAPAKK
: .::. . .
Mflv_1125|M.gilvum_PYR-GCK -PPADA-VTSAAKEAAAQAKSTVATASTAVTSPPPAPPGPASSSRLPLAA
Mvan_5681|M.vanbaalenii_PYR-1 APPVQASLAAAAEETAAQAKSTVATASTAVSAPPPAPPAGLDSSRLPIVA
Mb3882|M.bovis_AF2122/97 RIETNGQLAAAAKDAAAQAKSTVEGANDALARNASVPAPSHSPVPLIV--
Rv3852|M.tuberculosis_H37Rv RIETNGQLAAAAKDAAAQAKSTVEGANDALARNASVPAPSHSPVPLIV--
MLBr_00067|M.leprae_Br4923 RIETNGQLD-VAKDVAEQAQSAVEGASNPVPNGAEALEASNSPVALVI--
MAV_0176|M.avium_104 RADTNGDLTAAAKDAAAQAKSTVEAADNPVAPAVVGPAAGQSPVPLIV--
MUL_5024|M.ulcerans_Agy99 QPSANGQLAEAAKDAAAQAKSTVDRANNPLPRVADE-QPSHSTVPLVV--
TH_2451|M.thermoresistible__bu GAAPPADLTSAAKEVAAHAKSTVASAPNPVATPAPISRPDDSRLPFMI--
MSMEG_6438|M.smegmatis_MC2_155 TAAPVDEIAAAAEEVAAQAKSTVEAAPNPVSGPVPVPVEEHHSTRFPVGP
MAB_3292c|M.abscessus_ATCC_199 AVVKKAAPVKKAPVKKAVVKKAAPVKKAVTKAPAKKA-ATKAPAKKAATK
MMAR_3834|M.marinum_M AAAKKETSAKKAPAKKAPAKKAAAKKAPAKKATAKKAPAKKAPAKKVPES
* .:.:.
Mflv_1125|M.gilvum_PYR-GCK AAVAGVLALVVALLVARRNSDD----------------------------
Mvan_5681|M.vanbaalenii_PYR-1 ALVAGVLALVVILLAARRNNDR----------------------------
Mb3882|M.bovis_AF2122/97 AVTLSLLALLLIRQLRRR--------------------------------
Rv3852|M.tuberculosis_H37Rv AVTLSLLALLLIRQLRRR--------------------------------
MLBr_00067|M.leprae_Br4923 ALAIGLLALLLIHQLRRR--------------------------------
MAV_0176|M.avium_104 AVAVSLLAILLIRQLRRR--------------------------------
MUL_5024|M.ulcerans_Agy99 AIALSLLAMLLIRQLRRR--------------------------------
TH_2451|M.thermoresistible__bu AVASGALILLIVLLVNRRGK------------------------------
MSMEG_6438|M.smegmatis_MC2_155 TIAAAVMIVLLVLAARRRGAQD----------------------------
MAB_3292c|M.abscessus_ATCC_199 APAKKAPAKKAPAKKGRK--------------------------------
MMAR_3834|M.marinum_M AAAKAATPATDTELPAPKPVGEASNPTEIAPEPIAHDGDAQQLGAGRSDA
: . :
Mflv_1125|M.gilvum_PYR-GCK ------------
Mvan_5681|M.vanbaalenii_PYR-1 ------------
Mb3882|M.bovis_AF2122/97 ------------
Rv3852|M.tuberculosis_H37Rv ------------
MLBr_00067|M.leprae_Br4923 ------------
MAV_0176|M.avium_104 ------------
MUL_5024|M.ulcerans_Agy99 ------------
TH_2451|M.thermoresistible__bu ------------
MSMEG_6438|M.smegmatis_MC2_155 ------------
MAB_3292c|M.abscessus_ATCC_199 ------------
MMAR_3834|M.marinum_M AESSSSRPKGRP