For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ARIAYLGPEGTFTQAALLEIAAAGLVPGHDDGGAQPLPVESTPAALDAVRTGAAEFACVPIENSIDGSLA PTLDSLAIGSPLQVFAETTLDVAFSIVVRPGVGAADVRTLAAFPVAAAQVRQWLTAHLPSVELHPAYSNA DAARQVAEGQVDAAVTSPLAAAHWALQSLADGVVDESNARTRFLLIGVPGPPPPRTGTDRTSVVLRIANV PGALLDALTEFGMRGIDLTRIESRPTRTGLGTYMFFVDCVGHIADDAVAEALKALHRRCADVRYLGSWPT GQTYAGQPPPADEAAIWLQQLREGKPEASPGPPL*
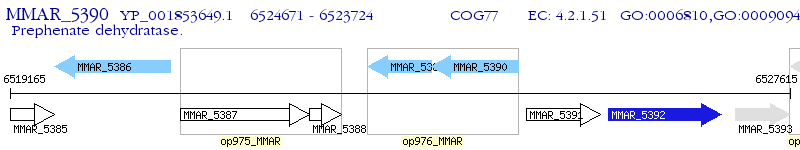
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5390 | pheA | - | 100% (315) | prephenate dehydratase PheA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3868c | pheA | 1e-134 | 77.60% (308) | prephenate dehydratase |
| M. gilvum PYR-GCK | Mflv_1145 | - | 1e-117 | 67.96% (309) | prephenate dehydratase |
| M. tuberculosis H37Rv | Rv3838c | pheA | 1e-134 | 77.60% (308) | prephenate dehydratase |
| M. leprae Br4923 | MLBr_00078 | pheA | 1e-133 | 75.56% (315) | prephenate dehydratase |
| M. abscessus ATCC 19977 | MAB_0132 | - | 1e-101 | 59.09% (308) | prephenate dehydratase |
| M. avium 104 | MAV_0188 | - | 1e-134 | 76.95% (308) | prephenate dehydratase |
| M. smegmatis MC2 155 | MSMEG_6418 | - | 1e-113 | 67.10% (307) | prephenate dehydratase |
| M. thermoresistible (build 8) | TH_1472 | pheA | 1e-114 | 65.62% (317) | POSSIBLE PREPHENATE DEHYDRATASE PHEA |
| M. ulcerans Agy99 | MUL_5011 | pheA | 1e-177 | 97.46% (315) | prephenate dehydratase |
| M. vanbaalenii PYR-1 | Mvan_5662 | - | 1e-113 | 66.35% (312) | prephenate dehydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1145|M.gilvum_PYR-GCK --MPGIAYLGPEGTFTEAALRALDAQGLIPATQ-----SGAGSVTPLATD
Mvan_5662|M.vanbaalenii_PYR-1 --MPGIAYLGPEGTFTEAALRALQAHGLIPSTAP--DAAGADEVTPIAAD
MMAR_5390|M.marinum_M --MARIAYLGPEGTFTQAALLEIAAAGLVP-------GHDDGGAQPLPVE
MUL_5011|M.ulcerans_Agy99 --MARIAYLGPEGTFTQAALLEIAAAGLVP-------GHDDGGAQPLPVD
Mb3868c|M.bovis_AF2122/97 --MVRIAYLGPEGTFTEAALVRMVAAGLVP-------ETGPDALQRMPVE
Rv3838c|M.tuberculosis_H37Rv --MVRIAYLGPEGTFTEAALVRMVAAGLVP-------ETGPDALQRMPVE
MLBr_00078|M.leprae_Br4923 MSVARIAYLGPEGTFTEAALLRMTAAGLVP-------DTGPDGLRRWPTE
MAV_0188|M.avium_104 --MARIAYLGPEGTFTEAALRQITAAGLVP-------GQGADGVRPTPVD
MSMEG_6418|M.smegmatis_MC2_155 --MPRIAYLGPEGTFTEAALLQMVAKGMVP-----GPAEDAGGFTPVRTD
TH_1472|M.thermoresistible__bu --MPRIAYLGPEGTFSESALLQMSARGMVPRAVPKDASGDPNGMTAVSVE
MAB_0132|M.abscessus_ATCC_1997 --MQRITYLGPEGTFSEAAMITLRTTGRIP---------GSSEVEPVSVA
: *:********:::*: : : * :* . . .
Mflv_1145|M.gilvum_PYR-GCK STPAALAAVRAGDADFACVPIENSIDGPVIPTLDSLADGVPLQIYAELTL
Mvan_5662|M.vanbaalenii_PYR-1 STSAALAAVRSGDADFACVPIENSIDGSVIPTLDSLADGAALQIYAELTL
MMAR_5390|M.marinum_M STPAALDAVRTGAAEFACVPIENSIDGSLAPTLDSLAIGSPLQVFAETTL
MUL_5011|M.ulcerans_Agy99 STPAALDAVRTGAAEFACVPIENSIDGSLAPTLDSLAIGSPLQVFAETTL
Mb3868c|M.bovis_AF2122/97 SAPAALAAVRDGGADYACVPIENSIDGSVLPTLDSLAIGVRLQVFAETTL
Rv3838c|M.tuberculosis_H37Rv SAPAALAAVRDGGADYACVPIENSIDGSVLPTLDSLAIGVRLQVFAETTL
MLBr_00078|M.leprae_Br4923 STPAALDAVRGGAADYACVPIENSIDGSVAPTLDNLAIGSPLQVFAETTL
MAV_0188|M.avium_104 GTPAALDAVRDGAADYACVPIENSIDGSVTPTLDSLAIGSPLQVFAETTL
MSMEG_6418|M.smegmatis_MC2_155 STPGALSAVREGRADYACVPIENSIDGTVLPTLDSLAAGSPLQIYAELTL
TH_1472|M.thermoresistible__bu STPAALAAVRSHDADYACVPIENSIEGSVLPTLDNLAVGNPLQIFAELTL
MAB_0132|M.abscessus_ATCC_1997 SAREALVQVQAGDADYACVPIESSLEGPVVPTLDTLAVGAPLQIFAETVL
.: ** *: *::******.*::*.: ****.** * **::** .*
Mflv_1145|M.gilvum_PYR-GCK DVSFTIAVRPGVTAADVRTVAAFPVAAAQVKRWLSENLPNVELVPSNSNA
Mvan_5662|M.vanbaalenii_PYR-1 DVSFTIAVRPGTAAADVRTVAAYPVAAAQVRRWLAAHLPEAEVVPANSNA
MMAR_5390|M.marinum_M DVAFSIVVRPGVGAADVRTLAAFPVAAAQVRQWLTAHLPSVELHPAYSNA
MUL_5011|M.ulcerans_Agy99 DVAFSIVVKPGVGAADVRTLAAFPVAAAQVRQWLTAHLPNVELHPAYSNA
Mb3868c|M.bovis_AF2122/97 DVTFSIVVKPGRNAADVRTLAAFPVAAAQVRQWLAAHLPAADLRPAYSNA
Rv3838c|M.tuberculosis_H37Rv DVTFSIVVKPGRNAADVRTLAAFPVAAAQVRQWLAAHLPAADLRPAYSNA
MLBr_00078|M.leprae_Br4923 DVEFNIVVKPGITAADIRTLAAFPVAAAQVRQWLAAHLAGAELRPAYSNA
MAV_0188|M.avium_104 DVAFSIVVKPGLSAADVRTLAAIGVAAAQVRQWVAANLAGAQLRPAYSNA
MSMEG_6418|M.smegmatis_MC2_155 DVAFTIVVRPGHDGP-VRTVAAFPVAAAQVRHWLAANLRDAEVVPAHSNA
TH_1472|M.thermoresistible__bu DVAFTIVTRPGTEPPDIATVAAFPVAAAQVRHWLADHLPHAQLVPATSNA
MAB_0132|M.abscessus_ATCC_1997 PVSFTIAVRPGTAAGDVKTVAGFPIAAAQVREWLATNLPDAELVAANSNA
* *.*..:** : *:*. :*****:.*:: :* .:: .: ***
Mflv_1145|M.gilvum_PYR-GCK AAARDVADGRAEAAVSTALATERYGLDTLAAGIVDEPNARTRFVLVGCPG
Mvan_5662|M.vanbaalenii_PYR-1 AAAQDVAAGRADAGVSTALATQRYGLEALAADVVDEPNARTRFVLVGRPG
MMAR_5390|M.marinum_M DAARQVAEGQVDAAVTSPLAAAHWALQSLADGVVDESNARTRFLLIGVPG
MUL_5011|M.ulcerans_Agy99 DGARQVAEGQVDAAVTSPLAAAHWALQSLADGVVDESNARTRFLLIGVPG
Mb3868c|M.bovis_AF2122/97 DAARQVADGLVDAAVTSPLAAARWGLAALADGVVDESNARTRFVLVGRPG
Rv3838c|M.tuberculosis_H37Rv DAARQVADGLVDAAVTSPLAAARWGLAALADGVVDESNARTRFVLVGRPG
MLBr_00078|M.leprae_Br4923 DAARQVAYGQVDAAVTSPLAATRWGLIALAAGIVDEPNARTRFVLVGMPG
MAV_0188|M.avium_104 DAAQQVAEGRADAAVTSPLAAARWGLDTLADGVVDEPNARTRFVLVGPPA
MSMEG_6418|M.smegmatis_MC2_155 AAAHDVAEGRADAGVSTRLAAERCGLDIMAADVVDEPNARTRFVLVGLPG
TH_1472|M.thermoresistible__bu AAAQDVAAGAADAGVSTALAAERYGLAVLATDVVDEPNARTRFVLVGQPA
MAB_0132|M.abscessus_ATCC_1997 AAAEDVKAERADAGVCTEWAAQRLGLHALASGVVDEAHAHTRFVLVGRPG
.*.:* .:*.* : *: : .* :* .:***.:*:***:*:* *.
Mflv_1145|M.gilvum_PYR-GCK PPPKRTGSDRTSVVLRLDNVPGALVTAMNELAIRGIDLTGIESRPTRTEL
Mvan_5662|M.vanbaalenii_PYR-1 PPPKCTGADRTSVVLQLDNVPGALVSAMTELAVRDIDLTRIESRPTRTGL
MMAR_5390|M.marinum_M PPPPRTGTDRTSVVLRIANVPGALLDALTEFGMRGIDLTRIESRPTRTGL
MUL_5011|M.ulcerans_Agy99 PPPPRTGTDRTSAVLRIANVPGALLDALTEFGMRGIDLTRIESRPTRTGL
Mb3868c|M.bovis_AF2122/97 PPPARTGADRTSAVLRIDNQPGALVAALAEFGIRGIDLTRIESRPTRTEL
Rv3838c|M.tuberculosis_H37Rv PPPARTGADRTSAVLRIDNQPGALVAALAEFGIRGIDLTRIESRPTRTEL
MLBr_00078|M.leprae_Br4923 PPPARTGTDRTSAVLRIDNAPGMLVAALAEFGIRGIDLTRIESRPTRTEL
MAV_0188|M.avium_104 PPPARTGADRTSVVLRIDNAPGALLAALAEFGIRGIDLTRIESRPTRTGL
MSMEG_6418|M.smegmatis_MC2_155 TPPPATGADRTAVVLRLVNEPGALVSAMTEFSIRDIDLTRIESRPTRTEL
TH_1472|M.thermoresistible__bu RPPRRTGADRTSVVLRLANVPGALVTALTELAVRDIDLTRIESRPTRTGL
MAB_0132|M.abscessus_ATCC_1997 PPPAATGADRTSVVLGLGNVPGALAAAMNEFAIRDIDLTRIESRPTRTGL
** **:***:.** : * ** * *: *:.:*.**** ******** *
Mflv_1145|M.gilvum_PYR-GCK GTYRFYLDFVGHIDDDAVAGALRALHRRCADVRYLGSWPTGETGG--AAP
Mvan_5662|M.vanbaalenii_PYR-1 GTYKFFLDFVGHIEDPPVAEALRALHRRCADVRYLGSWPTGDVVG--AAP
MMAR_5390|M.marinum_M GTYMFFVDCVGHIADDAVAEALKALHRRCADVRYLGSWPTGQTYA--GQP
MUL_5011|M.ulcerans_Agy99 GTYMFFIDCVGHIADDAVAEALKALHRRCADVRYLGSWPTGQTYA--AQP
Mb3868c|M.bovis_AF2122/97 GTYLFFVDCVGHIDDEAVAEALKAVHRRCADVRYLGSWPTGPAAG--AQP
Rv3838c|M.tuberculosis_H37Rv GTYLFFVDCVGHIDDEAVAEALKAVHRRCADVRYLGSWPTGPAAG--AQP
MLBr_00078|M.leprae_Br4923 GTYLFFVDCVGHIDDGVVAEALKALHRRCADVCYLGSWPAGLATGPTVSP
MAV_0188|M.avium_104 GIYRFFADCVGHIDDEPVAEALKALHRRCADVRYLGSWPTGTPAG--ALP
MSMEG_6418|M.smegmatis_MC2_155 GTYMFFLDCAGHIDDDPVAEALKALHRRCVDVRYLGSWPTESAAG--APP
TH_1472|M.thermoresistible__bu GSYMFFLDMVGHIDDDAVAGALMALHRRCADVRYLGSWPTGSVVG--APP
MAB_0132|M.abscessus_ATCC_1997 GTYRFFLDCVGHIDDIAVGEALKGLHRRCEDVRYLGSWPRGTTAPTGANP
* * *: * .*** * *. ** .:**** ** ****** *
Mflv_1145|M.gilvum_PYR-GCK PPLDEATAWLQRLREGRP--------------
Mvan_5662|M.vanbaalenii_PYR-1 PPMDESASWLEGLREGRP--------------
MMAR_5390|M.marinum_M PPADEAAIWLQQLREGKPEAS-----PGPPL-
MUL_5011|M.ulcerans_Agy99 PPADEAAIWLQQLREGKPEAS-----PEPPL-
Mb3868c|M.bovis_AF2122/97 PLVDEASRWLARLRAGKPEQTLVRPDDQGAQA
Rv3838c|M.tuberculosis_H37Rv PLVDEASRWLARLRAGKPEQTLVRPDDQGAQA
MLBr_00078|M.leprae_Br4923 PPPDEASRWLARLRAGKPDQ---ASEPGGGKL
MAV_0188|M.avium_104 PSTEEAVRWLAAVRDGKPEP------PGESRR
MSMEG_6418|M.smegmatis_MC2_155 PRLDEATTWLEGLRAG----------SGGA--
TH_1472|M.thermoresistible__bu PPLDEQARWLARLREGKPVSE-----TGGGP-
MAB_0132|M.abscessus_ATCC_1997 PVLDEASGWLAETREGRLR-------------
* :* ** * *