For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSEEVQSPGLAEAARDNLAAELARLRQRHDRLEIEVKNDRGMIGDHGDAAEAIQRADELAVLTERIHELD RRLKAGPSNATGSETLPSGTEVTLRFDDGEVVTMHVIAIVEENPVGHEYETLTAHSPLGQALAGRQAGDT VTYSTPRGPSQVELLSIKLPT
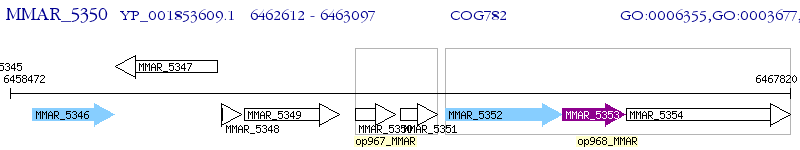
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5350 | - | - | 100% (161) | hypothetical protein MMAR_5350 |
| M. marinum M | MMAR_4387 | greA | 4e-05 | 23.13% (147) | transcription elongation factor GreA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3817 | - | 4e-70 | 79.50% (161) | nucleoside diphosphate kinase regulator |
| M. gilvum PYR-GCK | Mflv_2048 | greA | 2e-06 | 27.03% (148) | transcription elongation factor GreA |
| M. tuberculosis H37Rv | Rv3788 | - | 4e-70 | 79.50% (161) | nucleoside diphosphate kinase regulator |
| M. leprae Br4923 | MLBr_02393 | greA | 5e-07 | 25.00% (156) | transcription elongation factor GreA |
| M. abscessus ATCC 19977 | MAB_1201c | greA | 5e-07 | 27.39% (157) | transcription elongation factor GreA |
| M. avium 104 | MAV_0234 | - | 5e-63 | 75.62% (160) | nucleoside diphosphate kinase regulator |
| M. smegmatis MC2 155 | MSMEG_5263 | greA | 3e-07 | 27.70% (148) | transcription elongation factor GreA |
| M. thermoresistible (build 8) | TH_2169 | greA | 2e-09 | 28.66% (157) | PROBABLE TRANSCRIPTION ELONGATION FACTOR GREA (Transcript |
| M. ulcerans Agy99 | MUL_4967 | - | 3e-87 | 99.38% (161) | nucleoside diphosphate kinase regulator |
| M. vanbaalenii PYR-1 | Mvan_4668 | greA | 2e-06 | 26.35% (148) | transcription elongation factor GreA |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_5263|M.smegmatis_MC2_155 -MTDTQVTWLTQEAFDRLKAELDQLIANRPVIAAEINDRREEGDLRENGG
TH_2169|M.thermoresistible__bu -MTDTQTTWLTQEAYDRLKAELDQLIANRPIIAAEINDRREEGDLRENGG
Mflv_2048|M.gilvum_PYR-GCK -MTDTQVTWLTEEAYDRLKAELDQLIANRPVIAAEINDRREEGDLRENGG
Mvan_4668|M.vanbaalenii_PYR-1 -MTDTQVTWLTEESYDRLKAELDQLIANRPVIAAEINDRREEGDLRENGG
MLBr_02393|M.leprae_Br4923 -MTDTQVTWLTQESHDRLKAELDQLIANRPVIAAEINDRREEGDLRENGG
MAB_1201c|M.abscessus_ATCC_199 -MTDTQVTWLTQESHDRLKAELDQLIANRPVIAAEINERREEGDLRENGG
MMAR_5350|M.marinum_M MSEEVQSPGLAEAARDNLAAELARLRQRHDRLEIEVKNDR--GMIGDHGD
MUL_4967|M.ulcerans_Agy99 MSEEVQSPGLAEAARDNLAAELARLRQRHDRLEIEVKNDR--GMIGDHGD
Mb3817|M.bovis_AF2122/97 MSEKVESKGLADAARDHLAAELARLRQRRDRLEVEVKNDR--GMIGDHGD
Rv3788|M.tuberculosis_H37Rv MSEKVESKGLADAARDHLAAELARLRQRRDRLEVEVKNDR--GMIGDHGD
MAV_0234|M.avium_104 MTNKRQSP--ADAARKNLEAELDRLRQRRDRLEVEVKNDR--GMVGDHGD
. : :: : ..* *** :* .: : *::: * * : ::*.
MSMEG_5263|M.smegmatis_MC2_155 YHAAREEQGQQEARIRQLQELLNNAKVGEAPKQSGVALPGSVVKVYYDDD
TH_2169|M.thermoresistible__bu YHAAREEQGQQEARIRQLQELLNNAKVGEAPTVSGVALPGSVVTVYYDDD
Mflv_2048|M.gilvum_PYR-GCK YHAAREQQGQEEARIRQLQELLNDAKVGEAPKQSGIALPGSVVKVYYDDD
Mvan_4668|M.vanbaalenii_PYR-1 YHAAREQQGQEEARIRQLQELLNNAKVGEAPKQSGIALPGSVVKVYYDDD
MLBr_02393|M.leprae_Br4923 YHAAREEQGQQEARIRQLQDLLNIAKVGEAPKQSGVALPGSVVKVYYNDD
MAB_1201c|M.abscessus_ATCC_199 YHAAREEQGQQEARIRQLQELLNNAKVGEAPTQSGVALPGSVVKVYYDGD
MMAR_5350|M.marinum_M AAEAIQRADELAVLTERIHELDRRLKAGPSNATGSETLPSGTEVTLRFDD
MUL_4967|M.ulcerans_Agy99 AAEAIQRADELPVLTERIHELDRRLKAGPSNATGSETLPSGTEVTLRFDD
Mb3817|M.bovis_AF2122/97 AAEAIQRADELAILGDRINELDRRLRTGPTPWSGSETLPGGTEVTLRFPD
Rv3788|M.tuberculosis_H37Rv AAEAIQRADELAILGDRINELDRRLRTGPTPWSGSETLPGGTEVTLRFPD
MAV_0234|M.avium_104 AAEAIQRADELVVLSDRINELDRRLRAGPPDADTSATLPGGTEVTLRFAD
* :. .: ::::* . :.* . . :**... . *
MSMEG_5263|M.smegmatis_MC2_155 ENDTETFLIATRQEGISDGKLEVYSPNSPLGGALLDAKVGESRTYTVPSG
TH_2169|M.thermoresistible__bu ESDTETFLIGTRQEGITDGEVEVYSPNSPLGAALLDAKVGESRTYTVPSG
Mflv_2048|M.gilvum_PYR-GCK ENDTETFLIGTREQGVNDGKLEVYSPNSPLGGALIDAKVGESRSYTVPSG
Mvan_4668|M.vanbaalenii_PYR-1 EKDTETFLIGTREQGVSDGKLEVYSPNSPLGGALIDAKVGESRSYTVPSG
MLBr_02393|M.leprae_Br4923 KSDTETFLIATRQEGVNEGKLEVYSPNSPLGGALIDAKVGETRSYTVPNG
MAB_1201c|M.abscessus_ATCC_199 KGDTETFLIATRQEGVKDGKLEVYSPSSPLGGALIDAKVGETRSYTVPNG
MMAR_5350|M.marinum_M GEVVTMHVIAIVEENPVGHEYETLTAHSPLGQALAGRQAGDTVTYSTPRG
MUL_4967|M.ulcerans_Agy99 GEVVTMHVIAIVEENPVGHEYETLTAHSPLGQALAGRQAGDTVTYSTPRG
Mb3817|M.bovis_AF2122/97 GEVVTMHVISVVEETPVGREAETLTARSPLGQALAGHQPGDTVTYSTPQG
Rv3788|M.tuberculosis_H37Rv GEVVTMHVISVVEETPVGREAETLTARSPLGQALAGHQPGDTVTYSTPQG
MAV_0234|M.avium_104 GEVVTMHVISIVEETPVGREGETLTARSPLAQALAGRQPGDTVTYPTPQG
. .:*. :: : *. :. ***. ** . : *:: :*..* *
MSMEG_5263|M.smegmatis_MC2_155 NVVKVTLVSAEPYQG
TH_2169|M.thermoresistible__bu ATVKVTLVSAEPYQG
Mflv_2048|M.gilvum_PYR-GCK NTVKVTLVSAEPWHG
Mvan_4668|M.vanbaalenii_PYR-1 NTVKVTLVSAEPYHG
MLBr_02393|M.leprae_Br4923 NTVQVTLISAEPYHS
MAB_1201c|M.abscessus_ATCC_199 TEVQVTLVSAEPYHE
MMAR_5350|M.marinum_M -PSQVELLSIKLPT-
MUL_4967|M.ulcerans_Agy99 -PSQVELLSIKLPT-
Mb3817|M.bovis_AF2122/97 -PNQVQLLAVKLPS-
Rv3788|M.tuberculosis_H37Rv -PNQVQLLAVKLPS-
MAV_0234|M.avium_104 -EKQVELIAVKLP--
:* *:: :