For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTDTQVTWLTQEAFDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGYHAAREEQGQQEARIRQLQEL LNNAKVGEAPKQSGVALPGSVVKVYYDDDENDTETFLIATRQEGISDGKLEVYSPNSPLGGALLDAKVGE SRTYTVPSGNVVKVTLVSAEPYQG
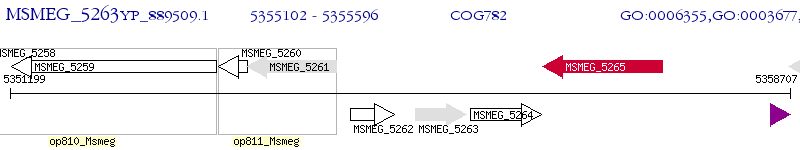
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_5263 | greA | - | 100% (164) | transcription elongation factor GreA |
| M. smegmatis MC2 155 | MSMEG_6292 | - | 4e-24 | 41.36% (162) | transcription elongation factor GreA |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1109c | greA | 3e-81 | 89.51% (162) | transcription elongation factor GreA |
| M. gilvum PYR-GCK | Mflv_2048 | greA | 2e-84 | 90.24% (164) | transcription elongation factor GreA |
| M. tuberculosis H37Rv | Rv1080c | greA | 3e-81 | 89.51% (162) | transcription elongation factor GreA |
| M. leprae Br4923 | MLBr_02393 | greA | 4e-82 | 89.51% (162) | transcription elongation factor GreA |
| M. abscessus ATCC 19977 | MAB_1201c | greA | 1e-80 | 89.51% (162) | transcription elongation factor GreA |
| M. marinum M | MMAR_4387 | greA | 6e-81 | 87.04% (162) | transcription elongation factor GreA |
| M. avium 104 | MAV_1204 | greA | 6e-79 | 89.81% (157) | transcription elongation factor GreA |
| M. thermoresistible (build 8) | TH_2169 | greA | 3e-83 | 91.46% (164) | PROBABLE TRANSCRIPTION ELONGATION FACTOR GREA (Transcript |
| M. ulcerans Agy99 | MUL_0200 | greA | 4e-81 | 87.04% (162) | transcription elongation factor GreA |
| M. vanbaalenii PYR-1 | Mvan_4668 | greA | 6e-85 | 90.85% (164) | transcription elongation factor GreA |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1109c|M.bovis_AF2122/97 MTDTQVTWLTQESHDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGY
Rv1080c|M.tuberculosis_H37Rv MTDTQVTWLTQESHDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGY
MAV_1204|M.avium_104 -----MTWLTQESHDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGY
MMAR_4387|M.marinum_M MTDTQVTWLTQESHDRLKAELDHLIANRPIIAAEINDRREEGDLRENGGY
MUL_0200|M.ulcerans_Agy99 MTDTQVTWLTQESHDRLKAELDHLIANRPIIAAEINDRREEGDLRENGGY
MLBr_02393|M.leprae_Br4923 MTDTQVTWLTQESHDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGY
MAB_1201c|M.abscessus_ATCC_199 MTDTQVTWLTQESHDRLKAELDQLIANRPVIAAEINERREEGDLRENGGY
MSMEG_5263|M.smegmatis_MC2_155 MTDTQVTWLTQEAFDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGY
TH_2169|M.thermoresistible__bu MTDTQTTWLTQEAYDRLKAELDQLIANRPIIAAEINDRREEGDLRENGGY
Mflv_2048|M.gilvum_PYR-GCK MTDTQVTWLTEEAYDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGY
Mvan_4668|M.vanbaalenii_PYR-1 MTDTQVTWLTEESYDRLKAELDQLIANRPVIAAEINDRREEGDLRENGGY
****:*:.********:******:******:*************
Mb1109c|M.bovis_AF2122/97 HAAREEQGQQEARIRQLQDLLSNAKVGEAPKQSGVALPGSVVKVYYNGDK
Rv1080c|M.tuberculosis_H37Rv HAAREEQGQQEARIRQLQDLLSNAKVGEAPKQSGVALPGSVVKVYYNGDK
MAV_1204|M.avium_104 HAAREEQGQQEARIRQLQDLLNNAKVGEAPKQSGVALPGSVVKVYYNGDK
MMAR_4387|M.marinum_M HAAREEQGQQEARIRQLQDLLNNAKVGEAPKQSGIALPGSVVKVYYNGDQ
MUL_0200|M.ulcerans_Agy99 HAAREEQGQQEARIRQLQDLLNNAKVGEAPKQSGIALPGSVVKVYYNGDQ
MLBr_02393|M.leprae_Br4923 HAAREEQGQQEARIRQLQDLLNIAKVGEAPKQSGVALPGSVVKVYYNDDK
MAB_1201c|M.abscessus_ATCC_199 HAAREEQGQQEARIRQLQELLNNAKVGEAPTQSGVALPGSVVKVYYDGDK
MSMEG_5263|M.smegmatis_MC2_155 HAAREEQGQQEARIRQLQELLNNAKVGEAPKQSGVALPGSVVKVYYDDDE
TH_2169|M.thermoresistible__bu HAAREEQGQQEARIRQLQELLNNAKVGEAPTVSGVALPGSVVTVYYDDDE
Mflv_2048|M.gilvum_PYR-GCK HAAREQQGQEEARIRQLQELLNDAKVGEAPKQSGIALPGSVVKVYYDDDE
Mvan_4668|M.vanbaalenii_PYR-1 HAAREQQGQEEARIRQLQELLNNAKVGEAPKQSGIALPGSVVKVYYDDDE
*****:***:********:**. *******. **:*******.***:.*:
Mb1109c|M.bovis_AF2122/97 SDSETFLIATRQEGVSDGKLEVYSPNSPLGGALIDAKVGETRSYTVPNGS
Rv1080c|M.tuberculosis_H37Rv SDSETFLIATRQEGVSDGKLEVYSPNSPLGGALIDAKVGETRSYTVPNGS
MAV_1204|M.avium_104 SDTETFLIATRQEGVNDGKLEVYSPNSPLGGALIDAKVGETRSYTVPNGS
MMAR_4387|M.marinum_M SDSETFLIATRQEGVNDGKLEVYSPNSPLGGALIDAKVGETRSYTVPNGS
MUL_0200|M.ulcerans_Agy99 SDSETFLIATRQEGVNDGKLEVYSPNSPLGGALIDAKVGETRSYTVPNGS
MLBr_02393|M.leprae_Br4923 SDTETFLIATRQEGVNEGKLEVYSPNSPLGGALIDAKVGETRSYTVPNGN
MAB_1201c|M.abscessus_ATCC_199 GDTETFLIATRQEGVKDGKLEVYSPSSPLGGALIDAKVGETRSYTVPNGT
MSMEG_5263|M.smegmatis_MC2_155 NDTETFLIATRQEGISDGKLEVYSPNSPLGGALLDAKVGESRTYTVPSGN
TH_2169|M.thermoresistible__bu SDTETFLIGTRQEGITDGEVEVYSPNSPLGAALLDAKVGESRTYTVPSGA
Mflv_2048|M.gilvum_PYR-GCK NDTETFLIGTREQGVNDGKLEVYSPNSPLGGALIDAKVGESRSYTVPSGN
Mvan_4668|M.vanbaalenii_PYR-1 KDTETFLIGTREQGVSDGKLEVYSPNSPLGGALIDAKVGESRSYTVPSGN
*:*****.**::*:.:*::*****.****.**:******:*:****.*
Mb1109c|M.bovis_AF2122/97 TVSVTLVSAEPYHS
Rv1080c|M.tuberculosis_H37Rv TVSVTLVSAEPYHS
MAV_1204|M.avium_104 TVEVTLVSAEPYHS
MMAR_4387|M.marinum_M VIEVTLISAEPYHS
MUL_0200|M.ulcerans_Agy99 VIEVTLISAEPYHS
MLBr_02393|M.leprae_Br4923 TVQVTLISAEPYHS
MAB_1201c|M.abscessus_ATCC_199 EVQVTLVSAEPYHE
MSMEG_5263|M.smegmatis_MC2_155 VVKVTLVSAEPYQG
TH_2169|M.thermoresistible__bu TVKVTLVSAEPYQG
Mflv_2048|M.gilvum_PYR-GCK TVKVTLVSAEPWHG
Mvan_4668|M.vanbaalenii_PYR-1 TVKVTLVSAEPYHG
:.***:****::