For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
GATGLSIGIVGATGQVGQVMRALLDERDFPATSVRFFASARSQGRKLPFRGQEIEVEDAETADPSGLDIA LFSAGGAMSKVQAPRFAAAGVTVIDNSSAWRKDPDVPLVVSEVNFERDVANRAGPLTKGIIANPNCTTMA AMPVLKVLHDEAGLVRIVVSSYQAVSGSGVAGVEELATQVREVIDGAERLVHDGAAVQFPAPVKYVAPIA FNVVPLAGSLVDDGSGETDEDQKLRFESRKILGIPDLSVSGTCVRVPVFTGHSLSINAEFERPLSPERAR ELLDGAPGVKVVDVPTPLAAAGVDESLVGRIRQDPGVPDGRGLALFVSGDNLRKGAALNTIQIAELLAAD R*
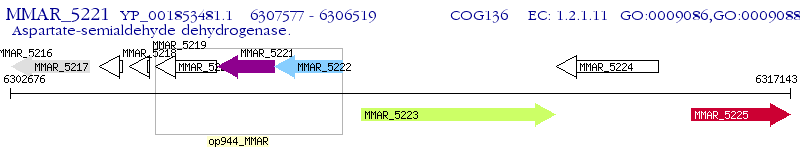
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5221 | asd | - | 100% (352) | aspartate-semialdehyde dehydrogenase Asd |
| M. marinum M | MMAR_2239 | gap | 3e-06 | 24.24% (231) | glyceraldehyde 3-phosphate dehydrogenase Gap |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3735c | asd | 1e-171 | 89.34% (347) | aspartate-semialdehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1305 | - | 1e-165 | 85.67% (342) | aspartate-semialdehyde dehydrogenase |
| M. tuberculosis H37Rv | Rv3708c | asd | 1e-171 | 89.34% (347) | aspartate-semialdehyde dehydrogenase |
| M. leprae Br4923 | MLBr_02322 | asd | 1e-165 | 84.81% (349) | aspartate-semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_0344 | - | 1e-161 | 84.30% (344) | aspartate-semialdehyde dehydrogenase |
| M. avium 104 | MAV_0394 | asd | 1e-172 | 89.57% (345) | aspartate-semialdehyde dehydrogenase |
| M. smegmatis MC2 155 | MSMEG_6256 | asd | 1e-160 | 82.37% (346) | aspartate-semialdehyde dehydrogenase |
| M. thermoresistible (build 8) | TH_2281 | asd | 1e-162 | 83.76% (351) | ASPARTATE-SEMIALDEHYDE DEHYDROGENASE ASD (ASA DEHYDROGENASE) |
| M. ulcerans Agy99 | MUL_4296 | asd | 0.0 | 98.30% (352) | aspartate-semialdehyde dehydrogenase |
| M. vanbaalenii PYR-1 | Mvan_5503 | - | 1e-161 | 84.84% (343) | aspartate-semialdehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5221|M.marinum_M MGATGLSIGIVGATGQVGQVMRALLDERDFPATSVRFFASARSQGRKLPF
MUL_4296|M.ulcerans_Agy99 MGATGLSIGIVGATGQVGQVMRVLLDERDFPATSVRFFASARSQGRKLPF
Mb3735c|M.bovis_AF2122/97 ---MGLSIGIVGATGQVGQVMRTLLDERDFPASAVRFFASARSQGRKLAF
Rv3708c|M.tuberculosis_H37Rv ---MGLSIGIVGATGQVGQVMRTLLDERDFPASAVRFFASARSQGRKLAF
MAV_0394|M.avium_104 ----MVAIGVVGATGQVGQVMRRLLDERDFPATSVRFFASSRSQGRKLEF
MLBr_02322|M.leprae_Br4923 MATRGLAIGVVGATGQVGHVMRTLLEQRDFRADSVRFFASARSQGRKLAF
Mflv_1305|M.gilvum_PYR-GCK ----MTRIGVVGATGQVGQVMRNLLVERDFPATEVRFFASARSAGKKLEF
Mvan_5503|M.vanbaalenii_PYR-1 ----MVRIGVVGATGQVGQVMRTLLAERDFPATGVRFFASARSAGKKLGF
MSMEG_6256|M.smegmatis_MC2_155 ----MVNIGVVGATGQVGQVMRNLLEQRNFPATSVRFFASPRSEGKKLTF
MAB_0344|M.abscessus_ATCC_1997 ----MTSIGVVGATGQVGAVMRKLLEEREFPASSVRFFASARSEGKKLAF
TH_2281|M.thermoresistible__bu --MGAVNLGVVGATGQVGQVMRALLEQRDFPVKSVRFFASARSQGKKLTF
:*:******** *** ** :*:* . ******.** *:** *
MMAR_5221|M.marinum_M RGQEIEVEDAETADPSGLDIALFSAGGAMSKVQAPRFAAAGVTVIDNSSA
MUL_4296|M.ulcerans_Agy99 RGQEIEVEDAETADPSGLDIALFSAGGAMSKVQAPRFAAAGVTVIDNSSA
Mb3735c|M.bovis_AF2122/97 RGQEIEVEDAETADPSGLDIALFSAGSAMSKVQAPRFAAAGVTVIDNSSA
Rv3708c|M.tuberculosis_H37Rv RGQEIEVEDAETADPSGLDIALFSAGSAMSKVQAPRFAAAGVTVIDNSSA
MAV_0394|M.avium_104 RGAEIEVEDAATADPTGLDIALFSAGKTMSLVQAPRFAAAGVTVIDNSSA
MLBr_02322|M.leprae_Br4923 RGQKIEVEDAETADPTGLDIALFSAGASMSRVQAPRFAAAGVTVIDNSSA
Mflv_1305|M.gilvum_PYR-GCK RGREIEVEDAETADPSGLDIALFSAGATMSRVQAPRFAAAGAVVVDNSSA
Mvan_5503|M.vanbaalenii_PYR-1 RGQEIEVEDASTADPSGLDIALFSAGATMSRVQAPRFAGAGAVVIDNSSA
MSMEG_6256|M.smegmatis_MC2_155 RGQEIEVENAETADPSGLDIALFSAGATMSRVQAPRFAEAGVIVVDNSSA
MAB_0344|M.abscessus_ATCC_1997 RGQEIEVENTETADPSGLDIALFSAGATMSRVQAPRFAQAGVTVIDNSSA
TH_2281|M.thermoresistible__bu RGQEIEVEDAATADPSGLDIALFSAGGAMSREQAPRFAAAGAVVIDNSSA
** :****:: ****:********** :** ****** **. *:*****
MMAR_5221|M.marinum_M WRKDPDVPLVVSEVNFERDVANRAGPLTKGIIANPNCTTMAAMPVLKVLH
MUL_4296|M.ulcerans_Agy99 WRKDPDVPLVVSEVNFERDVANRAGPLTKGIIANPNCTTMGAMSVLKVLH
Mb3735c|M.bovis_AF2122/97 WRKDPDVPLVVSEVNFERDAHRRP----KGIIANPNCTTMAAMPVLKVLH
Rv3708c|M.tuberculosis_H37Rv WRKDPDVPLVVSEVNFERDAHRRP----KGIIANPNCTTMAAMPVLKVLH
MAV_0394|M.avium_104 WRKDPDVPLVVSEVNFERDAHRRP----KGIIANPNCTTMAAMPVLKVLH
MLBr_02322|M.leprae_Br4923 WRRDAQVPLVVSEVNYNRDVRDAFP-LPKGIIANPNCTTMVSMPVLKVLH
Mflv_1305|M.gilvum_PYR-GCK WRKDPEVPLVVSEVNFERDVGNRHRALSKGIIANPNCTTMAAMPVLKPLH
Mvan_5503|M.vanbaalenii_PYR-1 WRKDPDVPLVVSEVNFERDAGNRP----KGIIANPNCTTMAAMPVLKPLH
MSMEG_6256|M.smegmatis_MC2_155 FRKDPDVPLVVSEVNFDRDVRGKK--LAKGIIANPNCTTMAAMPVLKPLH
MAB_0344|M.abscessus_ATCC_1997 WRKDPDVPLVVSEVNFDRDVRGVK--LPKGIIANPNCTTMAAMPVLKVLH
TH_2281|M.thermoresistible__bu WRKDPEVPLVVSEVNFDRDVRGRK--LAKNIIANPNCTTMAAMPVLKPLH
:*:*.:*********::**. *.********** :*.*** **
MMAR_5221|M.marinum_M DEAGLVRIVVSSYQAVSGSGVAGVEELATQVREVIDGAERLVHDGAAVQF
MUL_4296|M.ulcerans_Agy99 DEAGLVRIVVSSYQAVSGSGVAGVEELATQVREVIDGAEQLVHDGAALQF
Mb3735c|M.bovis_AF2122/97 DEARLVRLVVSSYQAVSGSGLAGVAELAEQARAVIGGAEQLVYDGGALEF
Rv3708c|M.tuberculosis_H37Rv DEARLVRLVVSSYQAVSGSGLAGVAELAEQARAVIGGAEQLVYDGGALEF
MAV_0394|M.avium_104 DEAGLQRLVVSSYQAVSGSGIAGVEELATQARAVIDGAEQLVHDGRAVSF
MLBr_02322|M.leprae_Br4923 DEAQLLRMVVSSYQAVSGSGLAGVAELAAQVRAVIDVAEQLIHDGGVVQF
Mflv_1305|M.gilvum_PYR-GCK DEAGLTRLIVSSYQAVSGSGLAGVEELATQARAVIDGAEKLVHDGSALDY
Mvan_5503|M.vanbaalenii_PYR-1 DEAGLTRLIVSSYQAVSGSGLAGVEELAGQARAVIDGVEQLVHDGSALQF
MSMEG_6256|M.smegmatis_MC2_155 EEAGLQRLIVSSYQAVSGSGIAGVEELAGQARAVIDGVEQLVHDGSALQY
MAB_0344|M.abscessus_ATCC_1997 DAAGLTRLIVSTYQAVSGSGLAGVEELATQVRAGIDGGEQLVHDGTAVTF
TH_2281|M.thermoresistible__bu DEAGLVRLIVSSYQAVSGSGLAGVEELYGQVRAVADDSRALVHDGSAVSF
: * * *::**:********:*** ** *.* . . *::** .: :
MMAR_5221|M.marinum_M PAPVKYVAPIAFNVVPLAGSLVDDGSGETDEDQKLRFESRKILGIPDLSV
MUL_4296|M.ulcerans_Agy99 PAPVKYVAPIAFNVVPLAGWLVDDGSGETDEDQKLRFESRKILGIPDLSV
Mb3735c|M.bovis_AF2122/97 PPPNTYVAPIAFNVVPLAGSLVDDGSGETDEDQKLRFESRKILGIPDLLV
Rv3708c|M.tuberculosis_H37Rv PPPNTYVAPIAFNVVPLAGSLVDDGSGETDEDQKLRFESRKILGIPDLLV
MAV_0394|M.avium_104 PAPQTYVAPIAFNVVPLAGSLVDDGSGETDEDQKLRHESRKILGVPDLAV
MLBr_02322|M.leprae_Br4923 PAPSKYVAPIAFNVVPLAGSLVDDDSGETDEDQKLRFESCKILGIPDLLV
Mflv_1305|M.gilvum_PYR-GCK PAPNKYVAPIAFNVVPLAGSLVDDGSGETDEDQKLRNESRKILGIPELLV
Mvan_5503|M.vanbaalenii_PYR-1 PPPNKYVAPIAFNVVPLAGSLVDDGSGETDEDQKLRNESRKILGIPELAV
MSMEG_6256|M.smegmatis_MC2_155 PAPNKYVAPIAFNIVPLAGNYVDDGSGETDEDQKLRNESRKILGIPELLV
MAB_0344|M.abscessus_ATCC_1997 PAPVKYVAPIAFNVIPLAGSLVDDGSGETDEDQKLRNESRKILGIPELLV
TH_2281|M.thermoresistible__bu PEPSTYVAPIAFNVVPLAGSMVDDGSGETDEDQKLRHESRKILDIPELAV
* * .********::**** ***.*********** ** ***.:*:* *
MMAR_5221|M.marinum_M SGTCVRVPVFTGHSLSINAEFERPLSPERARELLDGAPGVKVVDVPTPLA
MUL_4296|M.ulcerans_Agy99 SGTCVRVPVFTGHSLSINAEFERPLSPERARELLDGAPGVKVVDVPTPLA
Mb3735c|M.bovis_AF2122/97 SGTCVRVPVFTGHSLSINAEFAQPLSPERARELLDGATGVQLVDVPTPLA
Rv3708c|M.tuberculosis_H37Rv SGTCVRVPVFTGHSLSINAEFAQPLSPERARELLDGATGVQLVDVPTPLA
MAV_0394|M.avium_104 SGTCVRVPVFTGHSLSINAEFARPLSPERARELLDGAPGVKVVDVPTPLA
MLBr_02322|M.leprae_Br4923 SGTCVRVPVFTGHSLSINAEFARPLSPQRARDLLGGAPGVKLVDVPTPLA
Mflv_1305|M.gilvum_PYR-GCK SGTCVRVPVFTGHSLSINAEFSEPISVARATEILGGAPGVTLVDVPTPLA
Mvan_5503|M.vanbaalenii_PYR-1 SGTCVRVPVFTGHSLSINAEFSQPLSVARAKEILAGAAGVTLVDVPTPLA
MSMEG_6256|M.smegmatis_MC2_155 SGTCVRVPVFSGHSLSINAEFSQPISVERTKELLSAAAGVKLVDVPTPLA
MAB_0344|M.abscessus_ATCC_1997 SGTCVRVPVFTGHSLSINAEFERELTPARATELLSAAAGVTLADVPTPLA
TH_2281|M.thermoresistible__bu SGTCVRVPVYTGHSLSVNAEFARPLSVARATELLADAPGVKLVDVPTPLA
*********::*****:**** . :: *: ::* *.** :.*******
MMAR_5221|M.marinum_M AAGVDESLVGRIRQDPGVPDGRGLALFVSGDNLRKGAALNTIQIAELLAA
MUL_4296|M.ulcerans_Agy99 AAGVDESLVGRIRQDPGVPDGRGLALFVSGDNLRKGAALNTIQIAELLAA
Mb3735c|M.bovis_AF2122/97 AAGVDESLVGRIRRDPGVPDGRGLALFVSGDNLRKGAALNTIQIAELLTA
Rv3708c|M.tuberculosis_H37Rv AAGVDESLVGRIRRDPGVPDGRGLALFVSGDNLRKGAALNTIQIAELLTA
MAV_0394|M.avium_104 AAGVDESLVGRIRRDPGVPDGRGLALFISGDNLRKGAALNTIQIAELLAA
MLBr_02322|M.leprae_Br4923 SAGVDESLVGRIRRDPGVPDGRGLALFVSGDNLRKGAALNAIQIAELLAT
Mflv_1305|M.gilvum_PYR-GCK AAGADDSLVGRIRQDPGAPDGRGLALFVSGDNLRKGAALNTIQIAELLAR
Mvan_5503|M.vanbaalenii_PYR-1 AAGADDSLVGRIRRDPGVPDDRGLALFVSGDNLRKGAALNTIQIAELLAA
MSMEG_6256|M.smegmatis_MC2_155 AAGIDDCLVGRIRQDPGVPDGRGLALFVSGDNLRKGAALNTIQIAELLAA
MAB_0344|M.abscessus_ATCC_1997 AAGADDSLVGRIRQDPGVPGNRGLALFISGDNLRKGAALNTIQIAELLVA
TH_2281|M.thermoresistible__bu AAGVDESLVGRIRQDPGVPDGRGLALFVSGDNLRKGAALNTIQIAEMLAA
:** *:.******:***.*..******:************:*****:*.
MMAR_5221|M.marinum_M DR
MUL_4296|M.ulcerans_Agy99 DR
Mb3735c|M.bovis_AF2122/97 DL
Rv3708c|M.tuberculosis_H37Rv DL
MAV_0394|M.avium_104 QL
MLBr_02322|M.leprae_Br4923 QL
Mflv_1305|M.gilvum_PYR-GCK G-
Mvan_5503|M.vanbaalenii_PYR-1 QL
MSMEG_6256|M.smegmatis_MC2_155 DL
MAB_0344|M.abscessus_ATCC_1997 G-
TH_2281|M.thermoresistible__bu DL