For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NGLPRVGLGVDVHPIEPGRPCWLVGLLFPSADGCAGHSDGDVAAHALCDAVLSAAGLGDIGDVFGVGDPR WEGVSGATMLRHVVELITQHGYRVGNASVQVVGNRPKIGWRRYEAQAVLSRLLNAPVSVSATTTDGLGLT GRGEGLAAIATALVVSLRDPLGSQ*
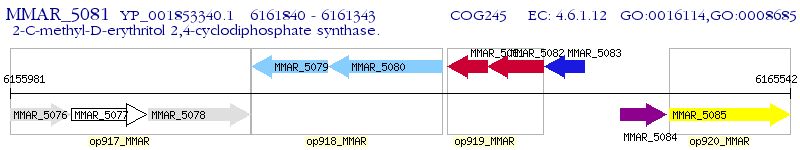
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5081 | ispF | - | 100% (165) | 2C-methyl-D-erythritol 2,4-cyclodiphosphate synthase IspF |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3612c | ispF | 2e-83 | 93.08% (159) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. gilvum PYR-GCK | Mflv_1445 | ispF | 2e-64 | 76.13% (155) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. tuberculosis H37Rv | Rv3581c | ispF | 2e-83 | 93.08% (159) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. leprae Br4923 | MLBr_00322 | ispF | 2e-70 | 80.38% (158) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. abscessus ATCC 19977 | MAB_0570 | - | 5e-66 | 72.78% (158) | putative 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. avium 104 | MAV_0572 | ispF | 4e-71 | 81.41% (156) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. smegmatis MC2 155 | MSMEG_6075 | ispF | 1e-62 | 72.50% (160) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. thermoresistible (build 8) | TH_1184 | - | 8e-69 | 76.92% (156) | PROBABLE 2C-METHYL-D-ERYTHRITOL 2,4-CYCLODIPHOSPHATE SYNTHASE |
| M. ulcerans Agy99 | MUL_4157 | ispF | 4e-90 | 100.00% (159) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
| M. vanbaalenii PYR-1 | Mvan_5339 | ispF | 1e-66 | 80.79% (151) | 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6075|M.smegmatis_MC2_155 --------MMLPRVGLGTDVHPIEAGRPCRLLCLEFDDADGCAGHSDGDV
TH_1184|M.thermoresistible__bu LPPHPDQVMSIPRVGLGTDVHPIEPGRPCWLLGLLFDGVDGCAGHSDGDV
MAB_0570|M.abscessus_ATCC_1997 -------MNALPRVGIGTDVHPIEEGKPCWLLGLLFSDENGCEGHSDGDV
Mflv_1445|M.gilvum_PYR-GCK --------MSTFRVGLGTDVHPIEAGRPCRLLCLLFDDGDGCAGHSDGDV
Mvan_5339|M.vanbaalenii_PYR-1 -------MLPSIRVGLGTDVHPIEAGRPCWLLCLLFDDADGCAGHSDGDV
MMAR_5081|M.marinum_M -------MNGLPRVGLGVDVHPIEPGRPCWLVGLLFPSADGCAGHSDGDV
MUL_4157|M.ulcerans_Agy99 -------------MGLGVDVHPIEPGRPCWLVGLLFPSADGCAGHSDGDV
Mb3612c|M.bovis_AF2122/97 -------MNQLPRVGLGTDVHPIEPGRPCWLVGLLFPSADGCAGHSDGDV
Rv3581c|M.tuberculosis_H37Rv -------MNQLPRVGLGTDVHPIEPGRPCWLVGLLFPSADGCAGHSDGDV
MLBr_00322|M.leprae_Br4923 -------MSELPRVGLGFDVHVFEPGRPCWLVGLLFPDNDGCAGHSDGDV
MAV_0572|M.avium_104 -------MNTTPRIGLGVDVHPIQPGRPCRLLGLLFDDADGCAGHSDGDV
:*:* *** :: *:** *: * * . :** *******
MSMEG_6075|M.smegmatis_MC2_155 AAHALCDALLSAAGLGDLGTIFGTDRPQWRGASGADMIRHVRGLVENAGF
TH_1184|M.thermoresistible__bu AAHALCDALLSAAGLGDLGEVFGTGRPEWQGVTGADMLRHVRGLVEAEGF
MAB_0570|M.abscessus_ATCC_1997 AAHALCDALLSAAGLGDVGAVFGTGRPEWTGVSGAKMLSHVRDLLHDNGF
Mflv_1445|M.gilvum_PYR-GCK AAHALCDALLSAAGMGDIGEVFGTGLPQWRDVSGADMLRHVHTLLVEQGF
Mvan_5339|M.vanbaalenii_PYR-1 AAHALCDALLSAAGMGDIGEVFGTGLPQWRGVSGADMLRHVHSMMTAEGL
MMAR_5081|M.marinum_M AAHALCDAVLSAAGLGDIGDVFGVGDPRWEGVSGATMLRHVVELITQHGY
MUL_4157|M.ulcerans_Agy99 AAHALCDAVLSAAGLGDIGDVFGVGDPRWEGVSGATMLRHVVELITQHGY
Mb3612c|M.bovis_AF2122/97 AVHALCDAVLSAAGLGDIGEVFGVDDPRWQGVSGADMLRHVVVLITQHGY
Rv3581c|M.tuberculosis_H37Rv AVHALCDAVLSAAGLGDIGEVFGVDDPRWQGVSGADMLRHVVVLITQHGY
MLBr_00322|M.leprae_Br4923 AVHALCDAVLSAAGQGDIGGLFAVGDPRWERVSGADMLRHVVELLLRHGY
MAV_0572|M.avium_104 GAHALCDAVLSAAGLGDVGAVFGVDDPRWAGVSGADMLRHVADLTARHGF
..******:***** **:* :*... *.* .:** *: ** : *
MSMEG_6075|M.smegmatis_MC2_155 VIGNATVQVIGNRPKVGPRREEAQQVLSELVGAPVSVSATTTDGLGLTGR
TH_1184|M.thermoresistible__bu RIGNAAVQVIGNRPKVGPRRAEAQRLLTELLGAPVSVSATTTDGLGLTGR
MAB_0570|M.abscessus_ATCC_1997 QIGNAAVQVIGNRPKVGPRRAEAQELLSGLIGAPVSVSATTTDGLGLTGR
Mflv_1445|M.gilvum_PYR-GCK QVGNAAVQVIGNRPKIGPRRAEAQRILSELLDAPVSVSATTTDGLGLTGR
Mvan_5339|M.vanbaalenii_PYR-1 RVGNAAVQVIGNRPKIGPRRAEAQRVLSGILGAPVSVSATTTDGLGLTGR
MMAR_5081|M.marinum_M RVGNASVQVVGNRPKIGWRRYEAQAVLSRLLNAPVSVSATTTDGLGLTGR
MUL_4157|M.ulcerans_Agy99 RVGNASVQVVGNRPKIGWRRYEAQAVLSRLLNAPVSVSATTTDGLGLTGR
Mb3612c|M.bovis_AF2122/97 RVGNAVVQVIGNRPKIGWRRLEAQAVLSRLLNAPVSVSATTTDGLGLTGR
Rv3581c|M.tuberculosis_H37Rv RVGNAVVQVIGNRPKIGWRRLEAQAVLSRLLNAPVSVSATTTDGLGLTGR
MLBr_00322|M.leprae_Br4923 EVVNASVQVIGNRPKIGPRRTEAQRLLSALLRAPVSVAATTTEGLGLTGR
MAV_0572|M.avium_104 RVGNAAVQVIGNRPKVGPRRAEAQRVLSELLGAPVSVAATTTDGLGLTGR
: ** ***:*****:* ** *** :*: :: *****:****:*******
MSMEG_6075|M.smegmatis_MC2_155 GEGLAAIATALVAAEPVGDAK------
TH_1184|M.thermoresistible__bu GEGLAAIATALLVGGAVGGENRPAVGR
MAB_0570|M.abscessus_ATCC_1997 GEGLAAVATALVTAL------------
Mflv_1445|M.gilvum_PYR-GCK GEGLAAIATALVLAPADA---------
Mvan_5339|M.vanbaalenii_PYR-1 GEGLAAIATALVVGDEGKG--------
MMAR_5081|M.marinum_M GEGLAAIATALVVSLRDPLGSQ-----
MUL_4157|M.ulcerans_Agy99 GEGLAAIATALVVSLRDPLGSQ-----
Mb3612c|M.bovis_AF2122/97 GEGLAAIATALVVSLR-----------
Rv3581c|M.tuberculosis_H37Rv GEGLAAIATALVVSLR-----------
MLBr_00322|M.leprae_Br4923 GEGLSAIATALVVQV------------
MAV_0572|M.avium_104 GEGLAAIATALVVPTG-----------
****:*:****: