For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSDRNVAVVGFAHAPHVRRTDGTTNGVEMLMPCFAQLYEDLGITQSDIGFWCSGSSDYLAGRAFSFISAI DSIGAVPPINESHVEMDAAWALFEAYIKILTGEVDTALVYGFGKSSAGILRQILSRQTDPYTVAPLWPDS VSMAGLQARLGLDSGKWTHEQMAQVALDSFARSQRNDSEKPAKSIDELLARPFFNDPLRRHDIAPITDGA AAIVLAADDRARELRESPAWITGFEHRIESPSLGVRDLTVSPSTAVSAKAASGGDTGIDVAEIHAPFTHQ HLIIAEAIGLTDKTKVNPSGGALAANPMFVAGLERIGFAARHIWDGSAGRVLAHATSGPALQQNLVAVME GNN
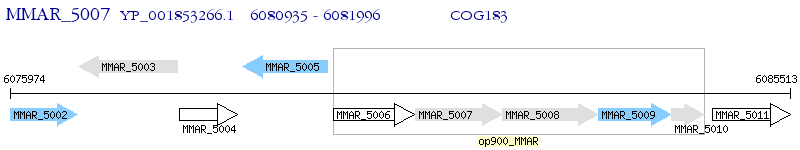
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5007 | ltp4 | - | 100% (353) | lipid transfer protein or keto acyl-CoA thiolase Ltp4 |
| M. marinum M | MMAR_5008 | ltp3 | 2e-11 | 25.00% (388) | lipid carrier protein or keto acyl-CoA thiolase Ltp3 |
| M. marinum M | MMAR_1918 | ltp1 | 7e-10 | 23.91% (297) | lipid-transfer protein Ltp1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3552 | - | 1e-175 | 85.23% (352) | lipid-transfer protein |
| M. gilvum PYR-GCK | Mflv_1537 | - | 1e-161 | 79.43% (350) | lipid-transfer protein |
| M. tuberculosis H37Rv | Rv3522 | ltp4 | 1e-174 | 84.94% (352) | lipid-transfer protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4168 | - | 1e-163 | 80.46% (348) | lipid-transfer protein |
| M. avium 104 | MAV_0637 | - | 1e-173 | 84.75% (354) | lipid-transfer protein |
| M. smegmatis MC2 155 | MSMEG_5922 | - | 1e-172 | 85.88% (347) | lipid-transfer protein |
| M. thermoresistible (build 8) | TH_2266 | - | 1e-170 | 83.52% (352) | POSSIBLE LIPID TRANSFER PROTEIN OR KETO ACYL-COA THIOLASE LTP4 |
| M. ulcerans Agy99 | MUL_4081 | ltp4 | 0.0 | 99.15% (353) | lipid-transfer protein |
| M. vanbaalenii PYR-1 | Mvan_5221 | - | 1e-167 | 82.57% (350) | lipid-transfer protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1537|M.gilvum_PYR-GCK MTD--VAVVGFAHAPHVRRTNGTTNGVEMLMPCFRELYSELGLEQTDIGF
Mvan_5221|M.vanbaalenii_PYR-1 MND--VAVVGFAHAPHVRRTDGTTNGVEMLMPCFASIYADLGITMSDIGF
MAB_4168|M.abscessus_ATCC_1997 MTD--VAVVGFAHAPHVRTTEGTTNGVEMLVPCFRQIYSELGITKSDIGF
TH_2266|M.thermoresistible__bu MTERSVAVVGFAHAPHVRRTDGTTNGVEMLMPCFAQLYDELGISQRDIGF
MSMEG_5922|M.smegmatis_MC2_155 -------MVGFAHAPHVRRTDGTTNGVEMLMPCFHQLYDELGLQQSDIGF
MMAR_5007|M.marinum_M MSDRNVAVVGFAHAPHVRRTDGTTNGVEMLMPCFAQLYEDLGITQSDIGF
MUL_4081|M.ulcerans_Agy99 MSDRNVAVVGFAHVPHVRRTDGTTNGVEMLMPCFAQLYEDLGITQSDIGF
Mb3552|M.bovis_AF2122/97 MSVRDIAVVGFAHAPHVRRTDGTTNGVEMLMPCFAQLYDELGITKADIGF
Rv3522|M.tuberculosis_H37Rv MSVRDIAVVGFAHAPHVRRTDGTTNGVEMLMPCFAQLYDELGITKADIGF
MAV_0637|M.avium_104 MSARNVAVVGFAHAPHVRRTDGTTNGVEMLMPCFAQLYQDLGITKADIGF
:*****.**** *:*********:*** .:* :**: ****
Mflv_1537|M.gilvum_PYR-GCK WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALYEAYIKIL
Mvan_5221|M.vanbaalenii_PYR-1 WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALYEAYIKIL
MAB_4168|M.abscessus_ATCC_1997 WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALYEAYIKIL
TH_2266|M.thermoresistible__bu WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALYEAYIKIL
MSMEG_5922|M.smegmatis_MC2_155 WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDGAWALYEAYIKIL
MMAR_5007|M.marinum_M WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALFEAYIKIL
MUL_4081|M.ulcerans_Agy99 WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALFEAYIKIL
Mb3552|M.bovis_AF2122/97 WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALYEAYIKLL
Rv3522|M.tuberculosis_H37Rv WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALYEAYIKLL
MAV_0637|M.avium_104 WCSGSSDYLAGRAFSFISAIDSIGAVPPINESHVEMDAAWALYEAYIKLL
*************************************.****:*****:*
Mflv_1537|M.gilvum_PYR-GCK TGEVETALVYGFGKSSAGTLRRVLALQTDPYTVAPLWPDSVSMAGLQARF
Mvan_5221|M.vanbaalenii_PYR-1 TGEVETALVYGFGKSSAGTLRRVLALQTDPYTVAPLWPDSVSMAGLQARF
MAB_4168|M.abscessus_ATCC_1997 SGQVETALVYGFGKSSAGILRRTLALQTDPYTVAPLWPDSVSLAALQARV
TH_2266|M.thermoresistible__bu TGEVDTALVYGFGKSSAGDLRRVLALQTDPYTVAPLWPDSVSIAGLQARL
MSMEG_5922|M.smegmatis_MC2_155 TGEVDTALVYGFGKSSAGVLRRVLALQTDPYSVAPLWPDAVSMAGLQARF
MMAR_5007|M.marinum_M TGEVDTALVYGFGKSSAGILRQILSRQTDPYTVAPLWPDSVSMAGLQARL
MUL_4081|M.ulcerans_Agy99 TGEVDTALVYGFGKSSAGILRQILSRQTDPYTVAPLWPDSVSMAGLQARL
Mb3552|M.bovis_AF2122/97 TGEVDTALVYGFGKSSAGTLRRVLSRQTDPYTVAPLWPDSVSMAGLQARL
Rv3522|M.tuberculosis_H37Rv TGEVDTALVYGFGKSSAGTLRRVLSRQTDPYTVAPLWPDSVSMAGLQARL
MAV_0637|M.avium_104 TGEVDTALVYGFGKSSAGVLRQILSRQTDPYTVAPLWPDSISMAGLQARM
:*:*:************* **: *: *****:*******::*:*.****.
Mflv_1537|M.gilvum_PYR-GCK GLDSGKWTAEQMARVALDALAASPRVDRLEPGNNVEELLDQPYFAEPLRR
Mvan_5221|M.vanbaalenii_PYR-1 GLDTGKWTAEQMAQVALDAQAAAPRVDSLEPGNSIEELLERPYFADPLRR
MAB_4168|M.abscessus_ATCC_1997 GLDSGKWTKEQMAQVALDAFRRAQRVDSEPSASSVDELLARDYFADPLHK
TH_2266|M.thermoresistible__bu GLDAGKWTAEQMARVALDAMAAAPRTDSVPPAETVDELLSRPYFADPLRR
MSMEG_5922|M.smegmatis_MC2_155 GLDAGKWTAEQMAQVALDAFAAGGRTDSVKPAKSIDELLERPFFADPLRR
MMAR_5007|M.marinum_M GLDSGKWTHEQMAQVALDSFARSQRNDSEKPAKSIDELLARPFFNDPLRR
MUL_4081|M.ulcerans_Agy99 GLDCGKWTHEQMAQVALDSFARSQRNDSEKPAKSIDELLARPFFNDPLRR
Mb3552|M.bovis_AF2122/97 GLDSGKWTHEQMARVAFDSFTNARRVDSVEPPITVGELLARPFFADPLRR
Rv3522|M.tuberculosis_H37Rv GLDSGKWTHEQMARVAFDSFTNARRVDSVEPPITVGELLARPFFADPLRR
MAV_0637|M.avium_104 GLDTGKWTEEQMARVAFDSFANARRVDNVEGSITVAELLERPFFAEPLRR
*** **** ****:**:*: . * * .: *** : :* :**::
Mflv_1537|M.gilvum_PYR-GCK HDIAPITDGASAIVLAAGDRARELRERPAWITGIEHRIETPVLGARDLTT
Mvan_5221|M.vanbaalenii_PYR-1 HDIAPITDGASAIVLASGDRARELRERPAWITGFEHRIETPVLGARDLTT
MAB_4168|M.abscessus_ATCC_1997 HDIAPISDGASIMVLAAGDRARELRENPAWISGIEHRVETPILGARDLAT
TH_2266|M.thermoresistible__bu HDIAPITDGASAIVLAAGDRARELRENPAWITGIEHRIETPVLGARDLTT
MSMEG_5922|M.smegmatis_MC2_155 HDIAPITDGAAAIVLAAGDRARELRENPAWITGIEHRIDTPVLGARDLTT
MMAR_5007|M.marinum_M HDIAPITDGAAAIVLAADDRARELRESPAWITGFEHRIESPSLGVRDLTV
MUL_4081|M.ulcerans_Agy99 HDIAPITDGAAAIVLAADDCARELRESPAWITGFEHRIESPSLGVRDLTV
Mb3552|M.bovis_AF2122/97 HDIAPITDGAAAVVLAADNRARELRENPAWITGIEHRIESPALGARDITE
Rv3522|M.tuberculosis_H37Rv HDIAPITDGAAAVVLAADNRARELRENPAWITGIEHRIESPALGARDITE
MAV_0637|M.avium_104 HDIAPITDGAAAIVLAAGDRARELRENPAWITGIEHRIESPALGVRDLTE
******:***: :***:.: ****** ****:*:***:::* **.**::
Mflv_1537|M.gilvum_PYR-GCK SPSTAASAAAATNGDPGSIEIAELYAPFSHQQLILTEAIGLGENTTINPS
Mvan_5221|M.vanbaalenii_PYR-1 SPSTAASAQAAAGGDTASIEIAELYAPFSHQQLILKEAIGLPDSTRINPS
MAB_4168|M.abscessus_ATCC_1997 SPSTAASAVAATGGDTGSIEVAELYAPFSHQQLILAEAIGLKDSTTVNPS
TH_2266|M.thermoresistible__bu SPSTAASAQAATGGDAGSIEVAELHAPFSHQQLILTEAIGLGSGTKVNPS
MSMEG_5922|M.smegmatis_MC2_155 SPSTAASAAAATGGDTASIEVAEIHAPFTHQHLILTEAIGLGGSTKINPS
MMAR_5007|M.marinum_M SPSTAVSAKAASGGDTG-IDVAEIHAPFTHQHLIIAEAIGLTDKTKVNPS
MUL_4081|M.ulcerans_Agy99 SPSTAVSAKAASGGDTG-IDVAEIHAPFTHQHLIIAEAIGLTDKTKVNPS
Mb3552|M.bovis_AF2122/97 SPSTKLAAKIATGGHTGDIDVAEIHGPFTHQHLIVAEAIRIPGKTKVNPS
Rv3522|M.tuberculosis_H37Rv SPSTKLAAKIATGGHTGDIDVAEIHGPFTHQHLIVAEAIRIPGKTKVNPS
MAV_0637|M.avium_104 SPSTKAAAEIATGGRTDNLDVVEICAPFTHQHLIIAEAIRIPGPTKVNPS
**** :* *:.* . :::.*: .**:**:**: *** : * :***
Mflv_1537|M.gilvum_PYR-GCK GGALAANPMFSAGLERIGLAARHIFEGNASRVLAHATSGPALQQNLVAVL
Mvan_5221|M.vanbaalenii_PYR-1 GGALAANPMFSAGLERIGFAARHIFDGNASRVLAHATSGPALQQNLVAVL
MAB_4168|M.abscessus_ATCC_1997 GGALVANPMFSAGLERIGFAAQHIWNGSAQRVLAHATSGPVLQQNLVAVL
TH_2266|M.thermoresistible__bu GGALSANPMFSAGLERIGFAARHIFDGSAQRVLAHATSGPALQQNLVAVL
MSMEG_5922|M.smegmatis_MC2_155 GGALAANPMFSAGLERIGFAARHIFDGSAGRVLAHATSGPALQQNLVAVL
MMAR_5007|M.marinum_M GGALAANPMFVAGLERIGFAARHIWDGSAGRVLAHATSGPALQQNLVAVM
MUL_4081|M.ulcerans_Agy99 GGALAANPMFVAGLERIGFAARHIWDGSAGRVLAHATSGPALQQNLVAVM
Mb3552|M.bovis_AF2122/97 GGPLAANPMFAAGLERIGFAAQHIWDGSARRVLAHATSGPALQQNLVAVM
Rv3522|M.tuberculosis_H37Rv GGPLAANPMFAAGLERIGFAAQHTWDGSARRVLAHATSGPALQQNLVAVM
MAV_0637|M.avium_104 GGALAANPMFVAGLERIGFAAQHIWDGSAERVLAHATSGPALQQNLVAVM
**.* ***** *******:**:* ::*.* **********.********:
Mflv_1537|M.gilvum_PYR-GCK EGKN
Mvan_5221|M.vanbaalenii_PYR-1 EGQS
MAB_4168|M.abscessus_ATCC_1997 EGK-
TH_2266|M.thermoresistible__bu EGK-
MSMEG_5922|M.smegmatis_MC2_155 EGKN
MMAR_5007|M.marinum_M EGNN
MUL_4081|M.ulcerans_Agy99 EGNN
Mb3552|M.bovis_AF2122/97 EGRG
Rv3522|M.tuberculosis_H37Rv EGRG
MAV_0637|M.avium_104 EGKN
**.