For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MPVSQHTIAGTVLTMPVRIRTANLHSAMFSVPAVAAQQLIDYSGLRVCEYLPGKAMVMQMLVRYVDGDLG RYHEYGTAVMVNPPGVQTRGPRALAKAAAFIHHLPVDQSFTLEAGRTIWGFPKIMADFNVREGKKFSFDV SADGQLIAGIEFGNGVAVPTRGAQVLQTYSHQDGVTREIPWEMRITGMRARLGGARLHLGDHPYARELAS LGLPKRALVSQSAANVEMTFGDAHAI
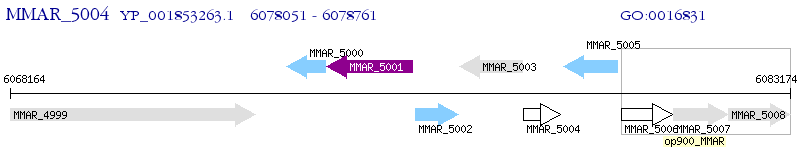
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_5004 | - | - | 100% (236) | hypothetical protein MMAR_5004 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3549 | - | 1e-118 | 85.59% (236) | hypothetical protein Mb3549 |
| M. gilvum PYR-GCK | Mflv_1540 | - | 3e-80 | 61.98% (242) | hypothetical protein Mflv_1540 |
| M. tuberculosis H37Rv | Rv3519 | - | 1e-118 | 85.59% (236) | hypothetical protein Rv3519 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. avium 104 | MAV_0640 | - | 2e-87 | 68.89% (225) | hypothetical protein MAV_0640 |
| M. smegmatis MC2 155 | MSMEG_5919 | - | 2e-88 | 67.93% (237) | hypothetical protein MSMEG_5919 |
| M. thermoresistible (build 8) | TH_2263 | - | 2e-46 | 65.19% (135) | HYPOTHETICAL PROTEIN Rv3519 |
| M. ulcerans Agy99 | MUL_4078 | - | 1e-133 | 98.31% (236) | hypothetical protein MUL_4078 |
| M. vanbaalenii PYR-1 | Mvan_5218 | - | 4e-82 | 63.87% (238) | hypothetical protein Mvan_5218 |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_5004|M.marinum_M MPVS--------QHTIAGTVLTMPVRIRTANLHSAMFSVPAVAAQQLIDY
MUL_4078|M.ulcerans_Agy99 MPVS--------QHIIAGTVLTMPVRIRTANLHSAMFSVPAVAAQQLIDY
Mb3549|M.bovis_AF2122/97 MPVS--------QHTIAGTVLTMPVRIRTANLHSAMFSVPADPAQRLIDY
Rv3519|M.tuberculosis_H37Rv MPVS--------QHTIAGTVLTMPVRIRTANLHSAMFSVPADPAQRLIDY
Mflv_1540|M.gilvum_PYR-GCK MSSARSRLCGVTQHTIAGTVLTMPVRVRRAYQHTAMFVVDAAAAQRMIDY
Mvan_5218|M.vanbaalenii_PYR-1 MTQVTS------QHTIAGTVLTMPVRVRTAHQHTAMFVVDADAAQRMIDY
MSMEG_5919|M.smegmatis_MC2_155 MT----------QHTIAGNTVTMPVRIRTADQHMAMFSVDADAAQRMIDH
MAV_0640|M.avium_104 ----------------------MPVRIRKADVHTAMFSVDAQAAQRLIDY
TH_2263|M.thermoresistible__bu -----------------------------------MFSVSADAAQRMIDY
** * * .**::**:
MMAR_5004|M.marinum_M SGLRVCEYLPG--KAMVMQMLVRYVDGDLGRYHEYGTAVMVNPPG---VQ
MUL_4078|M.ulcerans_Agy99 SGLRVCEYLPG--KAMVMQMLVRYVDGDLGRYHEYGTAVMVNPPG---VQ
Mb3549|M.bovis_AF2122/97 SGLRVCEYLPG--KAIVMQMLVRYVDGDLGRYHEYGTAIMVNPPG---TQ
Rv3519|M.tuberculosis_H37Rv SGLRVCEYLPG--KAIVMQMLVRYVDGDLGRYHEYGTAIMVNPPG---TQ
Mflv_1540|M.gilvum_PYR-GCK SGLRVCRFGRGNRRVLAVLMLMRYEDTDLGQYYEYGTNIMVNPPGTAPSQ
Mvan_5218|M.vanbaalenii_PYR-1 SGLRVCRFGRGHRKAMVVLMLMRYEDTDLGQYYEYGTNVMVNPPG---SN
MSMEG_5919|M.smegmatis_MC2_155 SGLQVCRYRPG--RAMVILMLMRYVDGDLGEYLEYGTNVMVNPPG---ST
MAV_0640|M.avium_104 SGLKVCQHRPG--RAVVNLMLARYIDGDLGQYHEFGTCVMVNPPG---ST
TH_2263|M.thermoresistible__bu SGLEVCQYRPG--KAVVNLMLARYFDGDLGQYHEFGTAVMVNPRG---SH
***.**.. * :.:. ** ** * ***.* *:** :**** *
MMAR_5004|M.marinum_M TRGPRALAKAAAFIHHLPVDQSFTLEAGRTIWGFPKIMADFNVREG-KKF
MUL_4078|M.ulcerans_Agy99 TRGPRALAKAAAFIHHLPVDQSFTLEAGRTIWGFPKIMADFNVREG-KKF
Mb3549|M.bovis_AF2122/97 RRGPRALTRAAAFIHHLPVDQVFTLEAGRTIWGFPKIMADFNVTDG-RRF
Rv3519|M.tuberculosis_H37Rv RRGPRALTRAAAFIHHLPVDQVFTLEAGRTIWGFPKIMADFNVTDG-RRF
Mflv_1540|M.gilvum_PYR-GCK ATGLRALQSAGAFVHHLPVDQSFTLEAGRTIWGYPKVMADFTIRHG-RQF
Mvan_5218|M.vanbaalenii_PYR-1 ATGMKALQSAGAFVHHLPVDQSFTLEAGRTIWGYPKVMADFTIRNG-RHF
MSMEG_5919|M.smegmatis_MC2_155 ATGLRALQSAGAYIHHLPVDQSFTLEAGRTIWGYPKVMADFTVREA-RQF
MAV_0640|M.avium_104 ARGWRALGEAAAFIHHLPVDQSFTLEAGRSIWGFPKIMADFTVREQ-GRF
TH_2263|M.thermoresistible__bu GHGLKAFGRAAAFIHHLPVDQDFTLEAGQKIWGFPKIMADFTVRDAGTLF
* :*: *.*::******* ******:.***:**:****.: . *
MMAR_5004|M.marinum_M SFDVSADGQLIAGIEFGNGVAVP---TRGAQVLQTYSHQDGVTREIPWEM
MUL_4078|M.ulcerans_Agy99 SFDVSADGQLIAGIEFSNGVAVP---TRGAQVLQTYSHQDGVTREIPWEM
Mb3549|M.bovis_AF2122/97 GFDVSADGRLIAGIEFSTGLPVP---TLGWQMLKTYSHHDGVTREIPWEM
Rv3519|M.tuberculosis_H37Rv GFDVSADGRLIAGIEFSTGLPVP---TLGWQMLKTYSHHDGVTREIPWEM
Mflv_1540|M.gilvum_PYR-GCK GFDVSIDGRHAVSMDFAPGIPVPSAFGSRPQVHSTYSHLDGVTRETEGQM
Mvan_5218|M.vanbaalenii_PYR-1 GFDVSIDGLHAVSMDFAPGIPVPAAFASKPQVHSTYSHLDGVTRETEGRM
MSMEG_5919|M.smegmatis_MC2_155 GFDVSIGGKFAVGMEFRPGIPVPPVFTSRPQVHPTYSHLDGVTRRTEGQM
MAV_0640|M.avium_104 GFDVSADGAHIAGIDFAPGLPVPSLFTSRPQVLKTYTFAEGTTREVPWEM
TH_2263|M.thermoresistible__bu GFDVREGDELIASMDFARGLPAP---ARRPPASP----------------
.*** .. ..::* *:..*
MMAR_5004|M.marinum_M RITGMRARLGGARLHLGDHPYARELASLGLPKRALVSQSAANVEMTFGDA
MUL_4078|M.ulcerans_Agy99 RITGMRARLGGARLHLGEHPYARELASLGLPKRALVSQSTANVEMTFGDA
Mb3549|M.bovis_AF2122/97 KVSGLRARLGGARLRLGDHPYAKELASLGLPKRALLSQSAANVEMTFGDG
Rv3519|M.tuberculosis_H37Rv KVSGLRARLGGARLRLGDHPYAKELASLGLPKRALLSQSAANVEMTFGDG
Mflv_1540|M.gilvum_PYR-GCK VLSGVRYRPGGVSLRLGEHPYAAELSSLGLPKRALVSSSAANVDMTFADA
Mvan_5218|M.vanbaalenii_PYR-1 VLSGVRYRPGGVSLRLGEHPYAAELSALGLPKRALVSSSAANVDMTFADS
MSMEG_5919|M.smegmatis_MC2_155 RLSGVRYRFGGVRLRLGDHPYAAELASLGLPKRALVSSSAANVQMTFGEA
MAV_0640|M.avium_104 RVRGLRGRPGGATLRLGTHPYAQELASLGLPTRAMISGSVGHVEMTFGDA
TH_2263|M.thermoresistible__bu --------PSRAPCRLTPSPTAPPAK---FPGR-----------------
. . :* * * :* *
MMAR_5004|M.marinum_M HAI---
MUL_4078|M.ulcerans_Agy99 HAI---
Mb3549|M.bovis_AF2122/97 HPI---
Rv3519|M.tuberculosis_H37Rv HPI---
Mflv_1540|M.gilvum_PYR-GCK KIVGE-
Mvan_5218|M.vanbaalenii_PYR-1 RTVGEA
MSMEG_5919|M.smegmatis_MC2_155 EVIS--
MAV_0640|M.avium_104 HPLG--
TH_2263|M.thermoresistible__bu ------