For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRRRVGRLPREPEIGASTHAAGESRLGDDGRPDAHPVRLDAELLMTTATCLMLPVMVTLDTTVVNVAQRT FIEEFSSTQAVVAWTSTGYTLSLASVIPLTGWAANRLGTKRLVMGSVLLFTLGSLLCAMASSITLLVAFR AAQGLGGGILIPLQLIILARAAGPARLGRVLTISMISVLMAPIAGPILGGWLIDAFGWQWIFLINLPIGA LTLILAGLVLPGDDSLPAESLDLVGMALLSPALVLLLYGLSALPARGTIGDPQVWLPTTVGLILIGGFAL HALHRGDHALIDLRLLKNRAVAAANATRFMFAVTFFGSCLLFPAYFQQVLGKTPLQSGLLLIPQTLAAAA VMPIVGRLMEKRGPRDVVLMGTALTVVGMGIFIYGMSREHVHLPALLIGLGTFGVGTAAMMVPVSWSAVH MLNSSEVAHGSTLFNVNHNVAASVGAALMSVVLTSRFNGSADIAAANRADEIREQALRQHLPLDVSNLPP RIRTPGFAEHLTKDLSLAYAGVFLLGMIVVATTAIPAWFLPKRPAPRIELLQVEP
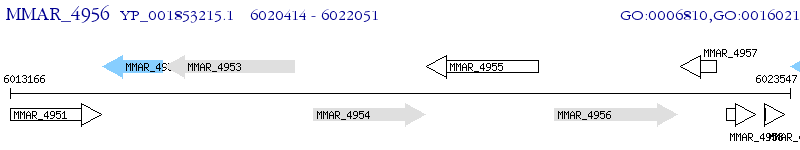
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4956 | - | - | 100% (545) | multidrug resistance integral membrane efflux protein |
| M. marinum M | MMAR_2515 | - | e-158 | 54.92% (508) | integral membrane drug efflux protein |
| M. marinum M | MMAR_4905 | - | e-149 | 52.01% (523) | integral membrane drug efflux protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0805c | emrB | 1e-142 | 49.91% (527) | multidrug resistance integral membrane efflux protein EmrB |
| M. gilvum PYR-GCK | Mflv_2221 | - | 6e-47 | 26.57% (493) | EmrB/QacA family drug resistance transporter |
| M. tuberculosis H37Rv | Rv0783c | emrB | 1e-142 | 49.91% (527) | multidrug resistance integral membrane efflux protein EmrB |
| M. leprae Br4923 | MLBr_01562 | - | 5e-32 | 26.15% (413) | putative transmembrane efflux protein |
| M. abscessus ATCC 19977 | MAB_1396 | - | 3e-47 | 27.35% (479) | EmrB/QacA family drug resistance transporter |
| M. avium 104 | MAV_0730 | - | 1e-150 | 51.57% (510) | EmrB efflux protein |
| M. smegmatis MC2 155 | MSMEG_5046 | - | 3e-44 | 28.94% (432) | drug transporter |
| M. thermoresistible (build 8) | TH_3965 | - | 1e-45 | 26.93% (479) | PUTATIVE drug transporter |
| M. ulcerans Agy99 | MUL_0542 | - | 0.0 | 99.75% (404) | multidrug resistance integral membrane efflux protein |
| M. vanbaalenii PYR-1 | Mvan_4474 | - | 5e-50 | 28.84% (423) | EmrB/QacA family drug resistance transporter |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4956|M.marinum_M --------------------MRRRVGRLPREPEIGASTHAAGESRLGDDG
MUL_0542|M.ulcerans_Agy99 --------------------------------------------------
Mb0805c|M.bovis_AF2122/97 ----------------------MLGNAMVEACPAEGDAPVPITPAGRPRS
Rv0783c|M.tuberculosis_H37Rv ----------------------MLGNAMVEACPAEGDAPVPITPAGRPRS
MAV_0730|M.avium_104 MRSPSAITELAAAGAGESGLFDMLSNAMDEAPYAAAKTP-PHAPTGQPGA
Mflv_2221|M.gilvum_PYR-GCK ----------------------------------MFDTVTSRAAGT----
Mvan_4474|M.vanbaalenii_PYR-1 ----------------------------------MFETVTARVRG-----
MSMEG_5046|M.smegmatis_MC2_155 ----------------------------------MPSSLNARIAGLLPSR
TH_3965|M.thermoresistible__bu ------------------------VTSVDAGRSTMSEVFTAYAPRS----
MAB_1396|M.abscessus_ATCC_1997 ----------------------------------MRPEYPR---------
MLBr_01562|M.leprae_Br4923 --------MVTLYDHERAVHNWTAARSDRPAPSRLTQQVEPASERTSKYP
MMAR_4956|M.marinum_M RPDAHPVRLDAELLMTTATCLMLPVMVTLDTTVVNVAQRTFIEEFSSTQA
MUL_0542|M.ulcerans_Agy99 --------------MTTATCLMLPVMVTLDTTVVNVAQRTFIEEFSSTQA
Mb0805c|M.bovis_AF2122/97 GQRSYPDRLDVGLLRTAGVCVLASVMAHVDVTVVSVAQRTFVADFGSTQA
Rv0783c|M.tuberculosis_H37Rv GQRSYPDRLDVGLLRTAGVCVLASVMAHVDVTVVSVAQRTFVADFGSTQA
MAV_0730|M.avium_104 TEREYPDKLDAALLRISGVCILATVMAILDVTVVSVAQRTFIDQFSSSQA
Mflv_2221|M.gilvum_PYR-GCK ----------TNPWPALWALLIGFFMILVDATIVAVANPAIMASLNAGYD
Mvan_4474|M.vanbaalenii_PYR-1 ----------ADPWPALWALLIGFFMILVDATIVPVANPAIMAQLGADYD
MSMEG_5046|M.smegmatis_MC2_155 VGALRPQDGTGNPWNALWAMMFGFFMILVDATIVSVANPTIMVELGADYD
TH_3965|M.thermoresistible__bu --------AAANPWHALWAMLIGFFMILVDATIVPVANLEIMADLGADYT
MAB_1396|M.abscessus_ATCC_1997 ------------PWITLWVVLFGLFMILLDSTIVSVANPAIKAGFGADYS
MLBr_01562|M.leprae_Br4923 TLLPSGRFPSGRFIAAVIVIGGMQLLATMDSTVAIVALPKIQNELSLSDA
. .: :* *:. ** : :.
MMAR_4956|M.marinum_M VVAWTSTGYTLSLASVIPLTGWAANRLGTKRLVMGSVLLFTLGSLLCAMA
MUL_0542|M.ulcerans_Agy99 VVAWTSTGYTLSLASVIPLTGWTANRLGTKRLVMGSVLLFTLGSLLCAMA
Mb0805c|M.bovis_AF2122/97 VVAWTMTGYMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCAVA
Rv0783c|M.tuberculosis_H37Rv VVAWTMTGYMLALATVIPTAGWAADRFGTRRLFMGSVLAFTLGSLLCAVA
MAV_0730|M.avium_104 VVAWTMTGYTLALATVIPITGWAADRFGTKRLFIGSVVAFMLGSLLCALA
Mflv_2221|M.gilvum_PYR-GCK AVIWVTSAYLLAYAVPLLVSGRLGDRYGPKNVYMVGLTVFTVASLWCGLS
Mvan_4474|M.vanbaalenii_PYR-1 AVIWVTSAYLLAYAVPLLVSGRLGDRYGPKTVYLVGLAVFTAASLWCGLS
MSMEG_5046|M.smegmatis_MC2_155 GVIWVTSAYLLAYAVPLLVTGRLGDMYGPKNLYLLGLGVFTAASLWCGLA
TH_3965|M.thermoresistible__bu AVIWVTSAYLLAYAVPLLIAGRLGDRFGPRTMYLIGLAIFTAASLWCGLA
MAB_1396|M.abscessus_ATCC_1997 STVWVTSAYLLGYAVPLLITGRLGDQFGPRSMYLSGLAVFTAASLWCGLA
MLBr_01562|M.leprae_Br4923 GRSWGITAYVLTFGGLMLLGGRLGDTIGRKRTFIVGVALFTISSVLCAVA
* :.* * . : * .: * : : .: * .*: *.::
MMAR_4956|M.marinum_M SSITLLVAFRAAQGLGGGILIPLQLIILARAAGP-ARLGRVLTISMISVL
MUL_0542|M.ulcerans_Agy99 SSITLLVAFRAAQGLGGGILIPLQLIILARAAGP-ARLGRVLTISMISVL
Mb0805c|M.bovis_AF2122/97 PNILLLIIFRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGIPML
Rv0783c|M.tuberculosis_H37Rv PNILLLIIFRVVQGFGGGMLTPVSFAILAREAGP-KRLGRVMAVVGIPML
MAV_0730|M.avium_104 ANILQLIVFRVVQGIGGGMLLPLGFMILTREAGP-RRLGRLMSILSIPML
Mflv_2221|M.gilvum_PYR-GCK DSIGMLIAARVVQGVGAALLTPQTMTVITRTFPA-HRRGVAMSVWGATAG
Mvan_4474|M.vanbaalenii_PYR-1 DSIGMLVAARVLQGVGAALLTPQTLTVITRTFPA-HNRGVAMSVWGATAG
MSMEG_5046|M.smegmatis_MC2_155 GSIEMLIAARVVQGIGAALLTPQTLSAITRIFPA-NRRGVAMSVWGATAG
TH_3965|M.thermoresistible__bu GTIGMLIAARVLQGLGAALLTPQTLSMITRIFPA-DRRGVPMGVWGATAG
MAB_1396|M.abscessus_ATCC_1997 GSIGWLIAARVVQGIGAAMLTPQTLTVVQRVFPP-ERRGTAMGVWGAVAG
MLBr_01562|M.leprae_Br4923 WDEVTLVIARLLQGIGSAIASPTGLALVATTFSKGSARNTATAVFAAMTA
*: * **.*..: * : : . :
MMAR_4956|M.marinum_M MAPIAGPILGGWLIDAFGWQWIFLINLPIGALTLILAGLVLPGDDSLPAE
MUL_0542|M.ulcerans_Agy99 MAPIAGPILGGWLIDAFGWQWIFLINLPIGALTLILAGLVLPGDDSLPAE
Mb0805c|M.bovis_AF2122/97 LGPVGGPILGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAASE
Rv0783c|M.tuberculosis_H37Rv LGPVGGPILGGWLIGAYGWRWIFLVNLPVGLSALVLAAIVFPRDRPAASE
MAV_0730|M.avium_104 LAPIAGPILGGWLIDTSSWRWIFLINVPIGLLTVALAAVVFPRDHPARSE
Mflv_2221|M.gilvum_PYR-GCK VATLVGPLAGGVLVDSLGWQWIFIVNVPVGIVGLVMALRLVPALPNPVRQ
Mvan_4474|M.vanbaalenii_PYR-1 VATLAGPLAGGVLVDSLGWQWIFIVNLPVGVLGFALAVWLVPSLPTARAQ
MSMEG_5046|M.smegmatis_MC2_155 VATLVGPLAGGVLVGGLGWQWIFFVNVPVGIVGIALAVWLVPTLPTTRHH
TH_3965|M.thermoresistible__bu VATLIGPLAGGVLVDGLGWQWIFIVNVPIGIAGLVLGARLMPRLP-RQDQ
MAB_1396|M.abscessus_ATCC_1997 IATLVGPVAGGLLVDGWGWQWIFYVNVPVGVLGIILGAIYIPRMPTHRHR
MLBr_01562|M.leprae_Br4923 IGSVMGLVVGGALTEVS-WRLAFLVNVPIGLVMIYLARTALHETHKERMK
:..: * : ** * *: * :*:*:* . :. . .
MMAR_4956|M.marinum_M SLDLVGMALLSPALVLLLYGLSALPARGTIGDPQVWLPTTVGLILIGGFA
MUL_0542|M.ulcerans_Agy99 SLDLVGMALLSPALVLLLYGLSALPARGTIGDPQVWLPTTVGLILIGGFA
Mb0805c|M.bovis_AF2122/97 NFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAAFV
Rv0783c|M.tuberculosis_H37Rv NFDYMGLLLLSPGLATFLFGVSSSPARGTMADRHVLIPAITGLALIAAFV
MAV_0730|M.avium_104 TFDAVGVLLLSPGLATFLFAVSSIPGRGTVADRHVLIPAAMGLTLIAGFV
Mflv_2221|M.gilvum_PYR-GCK GFDVPGVVLSGVGLFLIVFALQE--GQSHGWALWIWATVLAGVAVMAAFV
Mvan_4474|M.vanbaalenii_PYR-1 RFDLPGVVLSGVALFLIVFALQE--GQSHDWAPWIWATIGIGIAVMAAFL
MSMEG_5046|M.smegmatis_MC2_155 -FDIPGIVLSAVGMFLIVFALQE--GQSKDWAPWVWGTATLGVGVMAAFF
TH_3965|M.thermoresistible__bu GFDVLGMVLSAVGLFLIVFALQE--AQTHRWAPWIWGTLAGGAGVMAMFV
MAB_1396|M.abscessus_ATCC_1997 -LDLLGMALSAVGMFAFVFALQE--GESFQWAPGVWAMMAAGLVVLGVFV
MLBr_01562|M.leprae_Br4923 -LDAAGAILATLACTAAVFAFSMGPEKGWISLTTIGSGLVALVAAIAFAV
:* * * . ::... . : :.
MMAR_4956|M.marinum_M LHALHRGDHALIDLRLLKNRAVAAANATRFMFAVTFFGSCLLFPAYFQQV
MUL_0542|M.ulcerans_Agy99 LHALHRGDHALIDLRLLKNRAVAAANATRFMFAVTFFGSCLLFPAYFQQV
Mb0805c|M.bovis_AF2122/97 AHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQV
Rv0783c|M.tuberculosis_H37Rv AHSWYRTEHPLIDMRLFQNRAVAQANMTMTVLSLGLFGSFLLLPSYLQQV
MAV_0730|M.avium_104 GHAWHRADHPLIDLRLFRNPVLTHANVTMLVFATAFFGAGLLLPSYFQQV
Mflv_2221|M.gilvum_PYR-GCK YWQSINTAGALIPLVIFRDRNFSMSNLGVATIGFVATGMILPLMFFAQAV
Mvan_4474|M.vanbaalenii_PYR-1 YWQSVNTGEPLIPLIIFGDRNFSMSNLGVATIGFVATGMILPLMFYAQSV
MSMEG_5046|M.smegmatis_MC2_155 FWQSVNTREPLIPLRVFADRDFTLANIGVAVIGFAVTAMILPLMFYLQTV
TH_3965|M.thermoresistible__bu IWQSANRRHPLVPLQIFADRNFMLANVGVATIGFVVTAMVLPAMFFAQAV
MAB_1396|M.abscessus_ATCC_1997 WWQSANRGEPLIPLRLFADRNFSLAGVGIASMSFCVISTGLPMMFYTQLA
MLBr_01562|M.leprae_Br4923 VERTAEN--PVVPFDLFRDRNRLVTFIAIFLAGGVIFSLTVSIGLYIQDI
. .:: : :: : : . . : : *
MMAR_4956|M.marinum_M LGKTPLQSGLLLIPQTLAAAAVMPIVGRLMEKRGPRDVVLMGTALTVVGM
MUL_0542|M.ulcerans_Agy99 LGKTPLQSGLLLIPQTLAAAAVMPIVGRLMEKRGPRDVVLMGTALTVVGM
Mb0805c|M.bovis_AF2122/97 LHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLIAAGL
Rv0783c|M.tuberculosis_H37Rv LHQSPMQSGVHIIPQGLGAMLAMPIAGAMMDRRGPAKIVLVGIMLIAAGL
MAV_0730|M.avium_104 LHQTPMQAGVHMIPQGLGAMLTMRLTGPLVDRQGPGKVVLVGIALITAGL
Mflv_2221|M.gilvum_PYR-GCK CGLTPTRAALLTAPMAVATGVLAPFVGKLVDRAHPRSVVGFGFSLMAIGV
Mvan_4474|M.vanbaalenii_PYR-1 CGLTPTQAALLTAPMAVATGVLAPLVGKLVDRSHPRPVVGFGFALMAIGL
MSMEG_5046|M.smegmatis_MC2_155 CGLSPIRSALVTAPTAIASGVLAPFVGRIVDRVHPMPVIGFGFSAMAIGL
TH_3965|M.thermoresistible__bu CGLSPTGSALLIAPMAVASGVLAPMAGRIVDRSHPTPVVGFGFSVVAVGL
MAB_1396|M.abscessus_ATCC_1997 LGFSPTKAALTQAPTAIVSGILAPLAGWLVDRVRPSILIGGGIALMIAST
MLBr_01562|M.leprae_Br4923 LGYSALRAGIGFIPFVIAMGIGLGVSSQLVSRFSPRVLTIGGGWLLMVAM
:. :.: * : . . ::.: * : * .
MMAR_4956|M.marinum_M GIFIYGMSREHVHLPALLIGLGTFGVGTAAMMVPVSWSAVHMLNSSEVAH
MUL_0542|M.ulcerans_Agy99 GIFIYGMSREHVHLPALLIGLGTFGVGTAAMMVPVSWSAVHMLNSSEVAH
Mb0805c|M.bovis_AF2122/97 GTFAFGVARQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQIAR
Rv0783c|M.tuberculosis_H37Rv GTFAFGVARQADYLPILPTGLAIMGMGMGCSMMPLSGAAVQTLAPHQIAR
MAV_0730|M.avium_104 GAFAFGVARQAPYLPTLLAGLAITGLGMGCTMMPLSVASVQALAPHQIAR
Mflv_2221|M.gilvum_PYR-GCK TWLSIEMTPTTPIWRLLLP-LTAMGAAMAFIWSPLAATATRNLPPHLAGA
Mvan_4474|M.vanbaalenii_PYR-1 TWLSIEMTPSTPIWRLVVP-LTAMGVAMAFIWSPLAATATRNLPPQLAGA
MSMEG_5046|M.smegmatis_MC2_155 TWLSIEMAPTTPIWRLLLP-QLLMGIGMAFIWSPLAATATRHLPPDLAGA
TH_3965|M.thermoresistible__bu TWLAVEMAPTTPIWRLVLP-FVAVGVGMAFIWSPLATTATRQLPLHLSGA
MAB_1396|M.abscessus_ATCC_1997 LWWTLLMRPETQMWQLMLP-AAGIGAAMACIWGPLATTATRNLPPSVAGA
MLBr_01562|M.leprae_Br4923 LYGWAFMNRGVPYFPNLLAPIVVGGIGIGMAVVPLTLSAIAGVGFDRIGP
: : * . . *:: :: : .
MMAR_4956|M.marinum_M GSTLFNVNHNVAASVGAALMSVVLTSRFNGSADIAAANRADEIREQALRQ
MUL_0542|M.ulcerans_Agy99 GSTLFNVNHNVAASVGAALD--------------------------VGRT
Mb0805c|M.bovis_AF2122/97 GSTLISVNQQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGR
Rv0783c|M.tuberculosis_H37Rv GSTLISVNQQVGGSIGTALMSVLLTYQFNHSEIIATAKKVALTPESGAGR
MAV_0730|M.avium_104 GTTLMSVSHQVGGSMGTALMSMILTNQFNRSPNIVAANKLAALHQKAAAG
Mflv_2221|M.gilvum_PYR-GCK GSGVYNTTRQVGSVLGSAGMAALMSSELSTKIPG----ADAVPQSEGEVA
Mvan_4474|M.vanbaalenii_PYR-1 GSGVYNTTRQVGSVLGSAAMAALLASQLSAKIPG----EAPVP-VEGQAG
MSMEG_5046|M.smegmatis_MC2_155 GSGVYNTTRQVGSVLGSAGIAAFMTWRITDEVPGMQ-DAAPAGESGGAVE
TH_3965|M.thermoresistible__bu GSGVYNTTRQIGSVLGSAGMAAFLTGRLSAELPFG---ATAVP-AGEQTR
MAB_1396|M.abscessus_ATCC_1997 GSGVYNTLRQVGGVVGSSAMGALMTARLTATMPA-----MPPGSGSTPEA
MLBr_01562|M.leprae_Br4923 VSAIALMLQSLGGPLVLAVIQAVITSR-------------------TLYL
: : :.:.. : :
MMAR_4956|M.marinum_M HLPLDVSNLPPRIRTPGFAEHLTKDLSLAYAGVFLLGMIVVATTAIP---
MUL_0542|M.ulcerans_Agy99 HQPIQRQR------------------------------------------
Mb0805c|M.bovis_AF2122/97 GAAVDPSSLPRQ---TNFAAQLLHDLSHAYAVVFVIATALVVSTLIP---
Rv0783c|M.tuberculosis_H37Rv GAAVDPSSLPRQ---TNFAAQLLHDLSHAYAVVFVIATALVVSTLIP---
MAV_0730|M.avium_104 GTPIDQSAIPRQSLAPGFGGNVLHDLSHAYTAVFVIAVALVVCTIIP---
Mflv_2221|M.gilvum_PYR-GCK ALPQFLREPFALAMSQSMLLPAFVALFGVIAALFLFDLLGDRDAAAP--P
Mvan_4474|M.vanbaalenii_PYR-1 QLPAFLHAPFAAAMSQAMLLPAFVALFGVIAALFLVG-LGDRLPAPPRRP
MSMEG_5046|M.smegmatis_MC2_155 QLPEFLREPFSAAMSQAMLLPAFIALFGVVAALFLLGFGSNRAAGHP---
TH_3965|M.thermoresistible__bu VLPEFLHAPYAAAMAQSMLLPAFVALFGVLAAIFLAG----IVLSTP---
MAB_1396|M.abscessus_ATCC_1997 ALPPGLRAPFSAAMAQSMLLGIAALALGLVAALFMRP-GGSQVGAVP---
MLBr_01562|M.leprae_Br4923 GGTTGPVKFMNDAQLRALDHGYTYGLLWVAGAVVIVGVAALFIGYTP---
.
MMAR_4956|M.marinum_M --------------AWFLPKRPAPRIELLQVEP-----------------
MUL_0542|M.ulcerans_Agy99 --------------------------------------------------
Mb0805c|M.bovis_AF2122/97 --------------AAFLPKQQASHRRAPLLSA-----------------
Rv0783c|M.tuberculosis_H37Rv --------------AAFLPKQQASHRRAPLLSA-----------------
MAV_0730|M.avium_104 --------------ASFLPKKPAAETAGK---------------------
Mflv_2221|M.gilvum_PYR-GCK APDVEPLEPFDPDHPLDWADGEDDYVEYEVSREE---FDRLATDG-TADG
Mvan_4474|M.vanbaalenii_PYR-1 QETAEPVPAFDPDDELYWADGEDEYVEYEVPWDDGAEPRRPAQEADTVDA
MSMEG_5046|M.smegmatis_MC2_155 ----------EPEHAGDYPD-DDEYVEIILSSDP----------------
TH_3965|M.thermoresistible__bu ----------QPDADAQYLDEHDDYLVIELARPD----------------
MAB_1396|M.abscessus_ATCC_1997 ---------------------LDDVVSVED--------------------
MLBr_01562|M.leprae_Br4923 -------------EQVAYAQEVKEAVDAGEL-------------------
MMAR_4956|M.marinum_M --------------------------------------------------
MUL_0542|M.ulcerans_Agy99 --------------------------------------------------
Mb0805c|M.bovis_AF2122/97 --------------------------------------------------
Rv0783c|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0730|M.avium_104 --------------------------------------------------
Mflv_2221|M.gilvum_PYR-GCK AAYN-ITEDFTEDITD-----VLDTVVEAPPAPADSTPSQQWRDLVDQLL
Mvan_4474|M.vanbaalenii_PYR-1 AADTDVLDPVTEPMHDGGGYPAPESAPRRDHADPTSHPDREWRSILDQLL
MSMEG_5046|M.smegmatis_MC2_155 -----------------------------PEDPPTDPQDDRWQHILDRLM
TH_3965|M.thermoresistible__bu -------------------------PNPRTDIAVTAVIPALRPTLRPTLR
MAB_1396|M.abscessus_ATCC_1997 --------------------------------------------------
MLBr_01562|M.leprae_Br4923 --------------------------------------------------
MMAR_4956|M.marinum_M ------------------------------------
MUL_0542|M.ulcerans_Agy99 ------------------------------------
Mb0805c|M.bovis_AF2122/97 ------------------------------------
Rv0783c|M.tuberculosis_H37Rv ------------------------------------
MAV_0730|M.avium_104 ------------------------------------
Mflv_2221|M.gilvum_PYR-GCK DVSSPPGAP----RNGVGSADAEHPGGGPRV-----
Mvan_4474|M.vanbaalenii_PYR-1 DPPKPSDDPTGQGRNGFHVTPSEDGRGASRGRHSLD
MSMEG_5046|M.smegmatis_MC2_155 Q-DAPPEDP-------------LPGRGPQRR-----
TH_3965|M.thermoresistible__bu PSLGPPGGP----------VVQPNNTAALR------
MAB_1396|M.abscessus_ATCC_1997 ------------------------------------
MLBr_01562|M.leprae_Br4923 ------------------------------------