For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MQLTHFGHSCLLADFGQTRVLFDPGTFAHGFEGITGLAAILITHQHPDHVDVSRLPALLAANPDAALYAD AQTAAVLGTECREVHVGDELAVGELTIRAVGGRHAVIHPEIPVIENISYLVGDGDHRARLMHPGDALFVP DEPVDVLATPAAAPWMKISEAVDYLRSVAPARAVPIHQGIIATEARGIYYGRLAEMTDTDFQVLPEESAV TF
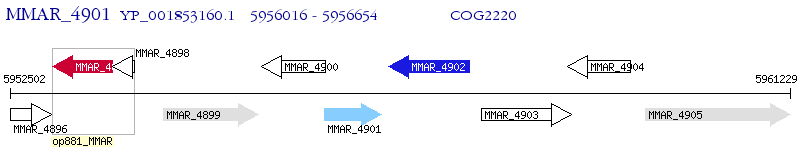
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4901 | - | - | 100% (212) | Zn-dependent hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0809c | - | 1e-61 | 87.60% (129) | hypothetical protein Mb0809c |
| M. gilvum PYR-GCK | Mflv_1622 | - | 3e-95 | 79.17% (216) | beta-lactamase domain-containing protein |
| M. tuberculosis H37Rv | Rv0786c | - | 1e-61 | 87.60% (129) | hypothetical protein Rv0786c |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0696c | - | 4e-88 | 70.83% (216) | hypothetical protein MAB_0696c |
| M. avium 104 | MAV_0733 | - | 1e-108 | 86.79% (212) | metallo-beta-lactamase superfamily protein, putative |
| M. smegmatis MC2 155 | MSMEG_5834 | - | 6e-93 | 76.44% (208) | metallo-beta-lactamase superfamily protein |
| M. thermoresistible (build 8) | TH_0565 | - | 1e-85 | 77.20% (193) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_5131 | - | 6e-92 | 74.65% (217) | beta-lactamase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1622|M.gilvum_PYR-GCK ---------------------------------------MQLTHFGHSCL
Mvan_5131|M.vanbaalenii_PYR-1 --------------------------------------MLQLTHFGHSCL
MSMEG_5834|M.smegmatis_MC2_155 MGAAPAGTATSVVAASPAAASNVAQINRMNPPTEGNNIEMQLTHFGHSCL
TH_0565|M.thermoresistible__bu --------------------------------------------------
Mb0809c|M.bovis_AF2122/97 --------------------------------------------------
Rv0786c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4901|M.marinum_M ---------------------------------------MQLTHFGHSCL
MAV_0733|M.avium_104 ---------------------------------------MQLTHFGHSCL
MAB_0696c|M.abscessus_ATCC_199 ---------------------------------------MQLTHFGHSCL
Mflv_1622|M.gilvum_PYR-GCK LASFT-GQGPDTSVLFDPGAFSHGFEGITGLAAILITHQHPDHVDVERLP
Mvan_5131|M.vanbaalenii_PYR-1 LASFDDGSGAQTTVLFDPGNFSHGFEGITGLDAILITHQHPDHADTARLP
MSMEG_5834|M.smegmatis_MC2_155 LATIS-----DTTVLFDPGNFSHGFEGITGLSAILITHQHPDHADTTRLP
TH_0565|M.thermoresistible__bu -------------VLFDPGIFSHGFEGITGLSAILITHQHPDHADTERLP
Mb0809c|M.bovis_AF2122/97 --------------------------------------------------
Rv0786c|M.tuberculosis_H37Rv --------------------------------------------------
MMAR_4901|M.marinum_M LADFG-----QTRVLFDPGTFAHGFEGITGLAAILITHQHPDHVDVSRLP
MAV_0733|M.avium_104 LAEFD-----HTRVLFDPGTFAHGFEGITGLTAILITHQHPDHIDATRLP
MAB_0696c|M.abscessus_ATCC_199 LAEFP-SENGNTTLLFDPGNFSHGFEGITGLSAILITHQHPDHVDLQRLP
Mflv_1622|M.gilvum_PYR-GCK ALIEANPQAALYADPQTAAQLGGPWQAVNVGDELSIGHLTVRGAGGTHAV
Mvan_5131|M.vanbaalenii_PYR-1 ALVEANPQAALFADPQTAAQLGGSWQAVNVGDQLSIGHLTVRGAGGTHAV
MSMEG_5834|M.smegmatis_MC2_155 ALLDANPQAKLYADPQTAAQLGEPWTAVHVGDEFTIGSINVRGVGGKHAV
TH_0565|M.thermoresistible__bu ALLEANPQAKLYADPQTAAQLGEPWQPVHVGDEFTIGHLTVRGAGGRHAV
Mb0809c|M.bovis_AF2122/97 ---------------------------MHVGDELPLAELTVRAVGGCHAV
Rv0786c|M.tuberculosis_H37Rv ---------------------------MHVGDELPLAELTVRAVGGCHAV
MMAR_4901|M.marinum_M ALLAANPDAALYADAQTAAVLGTECREVHVGDELAVGELTIRAVGGRHAV
MAV_0733|M.avium_104 ALLEGNPDAALYADPQTTAQLGAPCRAVHVGDELQIGELTVRAVGGKHAV
MAB_0696c|M.abscessus_ATCC_199 ALVEANPGAALYADPMTAAQLGGPWQAVHAGDNFTVADLTVRGVGGQHAV
::.**:: :. :.:*..** ***
Mflv_1622|M.gilvum_PYR-GCK IHPEIPVIDNISYLVGDGEHPARLMHPGDALFVPDEPVDVLATPAAAPWL
Mvan_5131|M.vanbaalenii_PYR-1 IHPEIPVIDNISYLVGDGEHPAKLMHPGDALFVPGEPVDVLATPAAAPWM
MSMEG_5834|M.smegmatis_MC2_155 IHPEIPVIDNISYLLGDDEHPARFMHPGDALFVPGEPVDVLATPAAAPWM
TH_0565|M.thermoresistible__bu IHPELPVIDNISYLIGDGEHPARLMHPGDALFTPGEPVDVLATPAAAPWL
Mb0809c|M.bovis_AF2122/97 IHPEIPVIENISYLVGDSKHRARLMHPGDALFVPGEQVDVLATPAAAPWM
Rv0786c|M.tuberculosis_H37Rv IHPEIPVIENISYLVGDSKHRARLMHPGDALFVPGEQVDVLATPAAAPWM
MMAR_4901|M.marinum_M IHPEIPVIENISYLVGDGDHRARLMHPGDALFVPDEPVDVLATPAAAPWM
MAV_0733|M.avium_104 IHPEIPVIENISFLVGDGGHRARLMHPGDALFVPDEPVDVLATPAAAPWM
MAB_0696c|M.abscessus_ATCC_199 IHPEIPLIDNTSYLIGDAEHAARLMHPGDALFAPGEDVDVLALPAAAPWM
****:*:*:* *:*:** * *::********.*.* ***** ******:
Mflv_1622|M.gilvum_PYR-GCK KISEAVDYLRAVAPTHAVPIHQAIIDPIGRPIFYGRLTEMTDTDFQVLEQ
Mvan_5131|M.vanbaalenii_PYR-1 KISEAIDYLRAVAPVHAVPIHQAIIDPIGRPIFYGRLTEMTDTDFRVLEQ
MSMEG_5834|M.smegmatis_MC2_155 KVSEAVDYLRAVAPRAAVPIHQGIIAPDARGVFYGRLSEMTSTDFQVLQE
TH_0565|M.thermoresistible__bu KISEAVDYLRAVAPNRAVPIHQAIIDPSARGIFYSRLQEMTDTDFQVLKE
Mb0809c|M.bovis_AF2122/97 KISEAVDYLRAVAPARAVPIHQAIVAPDARGIYYGRLTEMTTTDFQVLPE
Rv0786c|M.tuberculosis_H37Rv KISEAVDYLRAVAPARAVPIHQAIVAPDARGIYYGRLTEMTTTDFQVLPE
MMAR_4901|M.marinum_M KISEAVDYLRSVAPARAVPIHQGIIATEARGIYYGRLAEMTDTDFQVLPE
MAV_0733|M.avium_104 KVSEAVDYLRAVAPARAVPIHQGIIAPDARGIYYGRLSEMTSTDFQVLPE
MAB_0696c|M.abscessus_ATCC_199 KVSEAIEYLRAVNPARAVPIHQGILQEAAYPFYYTRFREMSDADFQVLPQ
*:***::***:* * ******.*: . .:* *: **: :**:** :
Mflv_1622|M.gilvum_PYR-GCK ENAVTF
Mvan_5131|M.vanbaalenii_PYR-1 ESAVTF
MSMEG_5834|M.smegmatis_MC2_155 ENGTEV
TH_0565|M.thermoresistible__bu ENATQF
Mb0809c|M.bovis_AF2122/97 ESAVTF
Rv0786c|M.tuberculosis_H37Rv ESAVTF
MMAR_4901|M.marinum_M ESAVTF
MAV_0733|M.avium_104 EDAVTF
MAB_0696c|M.abscessus_ATCC_199 EDAVTF
*... .