For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKHPPRSVVATAVTLAVVLAIGGCSSNGGSGGPGTATGTTVTTSSAEAATILKQAAEAMRKVTGMHVNLT VDGDVPNLRVTKLEGDISNSPQTVATGSATMLVGNKQEDAKFVYVDGHLYSDLGQPGTYTDFGDGASIYN VSVLLDPNQGLANLLANLKEASVTGSEQVDGVATTKITGNSAADDIATLAGSRLTSADVTTVPTTVWIAT DGSSHLVQIKILPTPATSVTLTMSDWGKQVTATKPV
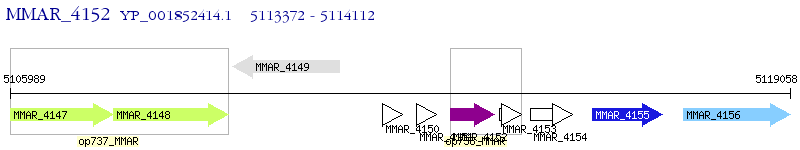
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4152 | lprA | - | 100% (246) | lipoprotein LprA |
| M. marinum M | MMAR_2220 | lprG | 7e-37 | 37.55% (237) | lipoprotein LprG |
| M. marinum M | MMAR_2188 | lprF | 4e-20 | 29.15% (271) | lipoprotein LprF |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1301c | lprA | 1e-109 | 81.38% (247) | putative lipoprotein LPRA |
| M. gilvum PYR-GCK | Mflv_3724 | - | 1e-39 | 34.85% (241) | hypothetical protein Mflv_3724 |
| M. tuberculosis H37Rv | Rv1270c | lprA | 1e-110 | 81.78% (247) | lipoprotein LprA |
| M. leprae Br4923 | MLBr_00557 | lprG | 4e-36 | 35.77% (246) | putative lipoprotein |
| M. abscessus ATCC 19977 | MAB_2806 | - | 2e-34 | 37.19% (199) | lipoprotein LprG precursor (27 kDa lipoprotein) |
| M. avium 104 | MAV_3367 | - | 3e-40 | 37.92% (240) | LprG protein |
| M. smegmatis MC2 155 | MSMEG_3070 | - | 7e-40 | 38.33% (240) | LprG protein |
| M. thermoresistible (build 8) | TH_1009 | lprG | 1e-41 | 37.30% (244) | PROBABLE CONSERVED LIPOPROTEIN LPRG |
| M. ulcerans Agy99 | MUL_4015 | lprA | 1e-136 | 99.19% (246) | lipoprotein LprA |
| M. vanbaalenii PYR-1 | Mvan_2692 | - | 3e-41 | 36.51% (241) | hypothetical protein Mvan_2692 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3724|M.gilvum_PYR-GCK MPPGRPAARSTRPDTRTLYDAGMQTR------------LLAIFAALFAAV
Mvan_2692|M.vanbaalenii_PYR-1 ----------------------MQTR------------LLAIFAALFAVV
MSMEG_3070|M.smegmatis_MC2_155 ----------------------MQTRP--------RFAVQSLFAILATAA
MLBr_00557|M.leprae_Br4923 ----------------------MQAPK-------HHRRLFAVLATLNTAT
MAV_3367|M.avium_104 ----------------------MQT----------RRRLSAVFASLTLAT
TH_1009|M.thermoresistible__bu ----------------------MQTRFRHAGRTVPVRNLFALFALVALIV
MAB_2806|M.abscessus_ATCC_1997 ----------------------MRNR------------IRLALIPVAVAA
MMAR_4152|M.marinum_M ----------------------MKHPP---------RSVVATA-VTLAVV
MUL_4015|M.ulcerans_Agy99 ----------------------MKHPP---------RSVVATA-VTLAVV
Mb1301c|M.bovis_AF2122/97 ----------------------MKHPP---------CSVVAAATAILAVV
Rv1270c|M.tuberculosis_H37Rv ----------------------MKHPP---------CSVVAAATAILAVV
*: : .
Mflv_3724|M.gilvum_PYR-GCK ALLAGCSS---------SSTEDSGKDLPDPTTLLSESSETTRNQTSAHLT
Mvan_2692|M.vanbaalenii_PYR-1 ALVAGCSG---------SSSKDSGKELPDAATLLQRSSETTKAQTSAHLN
MSMEG_3070|M.smegmatis_MC2_155 ALVAGCS----------SSSETSDAPLPDGAALLQESASKTKTQQSVHLL
MLBr_00557|M.leprae_Br4923 AVIAGCS----------SGSNLSSGPLPDATTWVKQATDITKNVTSAHLV
MAV_3367|M.avium_104 ALIAGCS----------SGSKQSGAPLPDPTSLVKQSADATKNVKSVHLV
TH_1009|M.thermoresistible__bu G-VAGCSSG-------EDQADQSEQPTPDAATLLEEAAQTTRDLQSVRLD
MAB_2806|M.abscessus_ATCC_1997 IALAGCS-----------KTDKADPNLPEAATLLSESAATTKTQTSTHIV
MMAR_4152|M.marinum_M LAIGGCSSNGGSGGPGTATGTTVTTSSAEAATILKQAAEAMRKVTGMHVN
MUL_4015|M.ulcerans_Agy99 LAIGGCSSNGGSGGPGTATGTTVTTSSAEAATILKQAAEAMRKVTGMHVN
Mb1301c|M.bovis_AF2122/97 LAIGGCSTEGDAG---KASDTAATASNGDAAMLLKQATDAMRKVTGMHVR
Rv1270c|M.tuberculosis_H37Rv LAIGGCSTEGDAG---KASDTAATASNGDAAMLLKQATDAMRKVTGMHVR
:.*** : : :..:: : . ::
Mflv_3724|M.gilvum_PYR-GCK LSVQGQIAELPVEQLEGDLTQTPAVAAKGTANIIFLGQRLNDVEFVVSDG
Mvan_2692|M.vanbaalenii_PYR-1 LSVQGQIPDLPVESLEGDLTQSPAVAAKGTANIIFLGQRLNDVAFVVSDG
MSMEG_3070|M.smegmatis_MC2_155 LTVQGKVDGLPVEKLDGDLTNAPAVAAEGTADLIAFGQKIADAKFVIADG
MLBr_00557|M.leprae_Br4923 LSVNGKITGLPVKTLTGDLTTHPNTVASGNATITLDGADLN-ANFVVVDG
MAV_3367|M.avium_104 LSIQGKISGLPIKTLTGDLTTTPATAAKGNATITLGGSDID-ANFVVVDG
TH_1009|M.thermoresistible__bu LTVQGEIPELPIETLQGDLTNVPEVGATGTADIIMMGQRFEDVDFVVAEG
MAB_2806|M.abscessus_ATCC_1997 LKVTGDKPTLKLSDLTGDLTTKPAVAAKGTAKT--GGLELP---FVVVDG
MMAR_4152|M.marinum_M LTVDGDVPNLRVTKLEGDISNSPQTVATGSATMLVGNKQED-AKFVYVDG
MUL_4015|M.ulcerans_Agy99 LTVDGDVPNLRVTKLEGDIANSPQTVATGSATMLVGNKQED-AKFVYVDG
Mb1301c|M.bovis_AF2122/97 LAVTGDVPNLRVTKLEGDISNTPQTVATGSATLLVGNKSED-AKFVYVDG
Rv1270c|M.tuberculosis_H37Rv LAVTGDVPNLRVTKLEGDISNTPQTVATGSATLLVGNKSED-AKFVYVDG
* : *. * : * **:: * . * *.* . ** :*
Mflv_3724|M.gilvum_PYR-GCK DLWATITAGGSLSNFGPAADVYDVAAILDPNVGLANVLANFSN-PSADGR
Mvan_2692|M.vanbaalenii_PYR-1 DLYAAITAGGALSNFGPASDVYDVAAILNPDVGLANVLANFSN-PTADGR
MSMEG_3070|M.smegmatis_MC2_155 NLYAALTPGDPLSNYGAAANIYDVSAILNPDTGLANVLANFSD-ATADGR
MLBr_00557|M.leprae_Br4923 ELYATLTPS-KWSDFGKASDIYDVASILNPDAGLANVLANFTG-AKTEGR
MAV_3367|M.avium_104 TLYATLTPN-KWSDFGKASDIYDVSVLLNPDNGLGNALANFSN-AKAEGR
TH_1009|M.thermoresistible__bu NLFAALAAG-DYIDFGPAADIYDVSAILNPDTGLANLLANFSDDATVEGR
MAB_2806|M.abscessus_ATCC_1997 ELFAQLGSA--YSSMGPVKDVYDVGLILDPNKGLANLLANITG-AKSEKT
MMAR_4152|M.marinum_M HLYSDLGQPGTYTDFGDGASIYNVSVLLDPNQGLANLLANLKE-ASVTGS
MUL_4015|M.ulcerans_Agy99 HLYSDLGQPGTYTDFGDGASIYNVSVLLDPNQGLANLLANLKE-ASVTGS
Mb1301c|M.bovis_AF2122/97 HLYSDLGQPGTYTDFGNGTSIYNVSVLLDPNKGLANLLANLKD-ASVAGS
Rv1270c|M.tuberculosis_H37Rv HLYSDLGQPGTYTDFGNGASIYNVSVLLDPNKGLANLLANLKD-ASVAGS
*:: : . * .:*:*. :*:*: **.* ***:. ..
Mflv_3724|M.gilvum_PYR-GCK ETVEGTQTVRVTGEVSADAVNKIAP---QIGATGPVPGTAWVAEEGDHKL
Mvan_2692|M.vanbaalenii_PYR-1 EEVKGAQTVRVTGEVSADAVNKIAP---QIAATGPVPGTAWITEDGDHEL
MSMEG_3070|M.smegmatis_MC2_155 ESINGTEAVRVKGNVSADAVNKIAP---ALKADGPVPGTAWITED-DHTL
MLBr_00557|M.leprae_Br4923 DSINGQSAVRISGNVSADAVNKIAP---PFNATQPMPATVWIQETGDHQL
MAV_3367|M.avium_104 ETINGQSTIRISGNVSADAVNKIMP---QFNATQPVPSTVWVQETGDHQL
TH_1009|M.thermoresistible__bu ETINGVRTLHVTGTVTPEAVNAIAP---QIGAEAPVPAGAWIRADGARDL
MAB_2806|M.abscessus_ATCC_1997 ETIDGVDSVLVTGTMSKDALNTFTG---GTTLTADIPAKAWIQKDGNHAL
MMAR_4152|M.marinum_M EQVDGVATTKITGNSAADDIATLAGSRLTSADVTTVPTTVWIATDGSSHL
MUL_4015|M.ulcerans_Agy99 EQVNGVATTKITGNSAADDIATLAGSRLTSADVTTVPTTVWIATDGSSHL
Mb1301c|M.bovis_AF2122/97 QQADGVATTKITGNSSADDIATLAGSRLTSEDVKTVPTTVWIASDGSSHL
Rv1270c|M.tuberculosis_H37Rv QQADGVATTKITGNSSADDIATLAGSRLTSEDVKTVPTTVWIASDGSSHL
: .* : :.* : : : : :* .*: *
Mflv_3724|M.gilvum_PYR-GCK MQVRLE-PSPGNSVTMTLSKWGEPVTVDKPAV--
Mvan_2692|M.vanbaalenii_PYR-1 MQARLE-PSPGNSVTMTLSQWGEPVTVDKPAV--
MSMEG_3070|M.smegmatis_MC2_155 LQAQLE-PTPGNSVTMTLSDWGKQVNVTKPAA--
MLBr_00557|M.leprae_Br4923 AQIRIDNKSSGNSVQMTLSNWDEPVQVTKPQVS-
MAV_3367|M.avium_104 VQANLQ-KSSGNSVQVTLSNWGEQVQVTKPPVSS
TH_1009|M.thermoresistible__bu VQAQLE-PDSGTTVTMTLSEWNKPVTVVKPEV--
MAB_2806|M.abscessus_ATCC_1997 TKISVD-TSPGNTIEMSLSDWGKPVTVDKPAQ--
MMAR_4152|M.marinum_M VQIKIL-PTPATSVTLTMSDWGKQVTATKPV---
MUL_4015|M.ulcerans_Agy99 VQIKIL-PTPATSVTLTMSDWGKQVTATKPV---
Mb1301c|M.bovis_AF2122/97 VQIQIA-PTKDTSVTLTMSDWGKQVTATKPV---
Rv1270c|M.tuberculosis_H37Rv VQIQIA-PTKDTSVTLTMSDWGKQVTATKPV---
: : .:: :::*.*.: * . **