For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DDVNSIVLAAGQAAEEGGTNNFLVPNGTFFFVLAIFLVVLAVIGTFVVPPILKVLRERDAMVAKTLADNK KSAEQFAAAQADYEKAMAEARVQASSYRDNARAEGRKVVEDARAHAEQEVASTLQQANEQLKRERDAVEL DLRANVGAMSATLANRIVGVDVTTPAAAG*
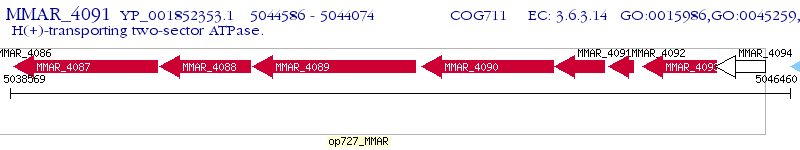
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_4091 | atpF | - | 100% (170) | ATP synthase B chain AtpF |
| M. marinum M | MMAR_4090 | atpH | 7e-10 | 24.46% (139) | ATP synthase delta chain AtpH |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1338 | atpF | 6e-72 | 82.84% (169) | F0F1 ATP synthase subunit B |
| M. gilvum PYR-GCK | Mflv_2314 | - | 1e-47 | 59.28% (167) | F0F1 ATP synthase subunit B |
| M. tuberculosis H37Rv | Rv1306 | atpF | 6e-72 | 82.84% (169) | F0F1 ATP synthase subunit B |
| M. leprae Br4923 | MLBr_01141 | atpF | 4e-63 | 75.00% (164) | F0F1 ATP synthase subunit B |
| M. abscessus ATCC 19977 | MAB_1449 | - | 1e-44 | 53.29% (167) | F0F1 ATP synthase subunit B |
| M. avium 104 | MAV_1523 | - | 3e-68 | 81.07% (169) | F0F1 ATP synthase subunit B |
| M. smegmatis MC2 155 | MSMEG_4940 | - | 2e-50 | 62.65% (166) | F0F1 ATP synthase subunit B |
| M. thermoresistible (build 8) | TH_1562 | - | 3e-42 | 56.00% (150) | bacteriophage lysis protein |
| M. ulcerans Agy99 | MUL_3958 | atpF | 5e-88 | 98.82% (170) | F0F1 ATP synthase subunit B |
| M. vanbaalenii PYR-1 | Mvan_4332 | - | 6e-47 | 60.53% (152) | F0F1 ATP synthase subunit B |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2314|M.gilvum_PYR-GCK -MGDLTSTN---LASAILAAEEGGGTSNFLLPNGTFFAVLLIFLIVLGVI
Mvan_4332|M.vanbaalenii_PYR-1 -MGELTTS--------IVAAEEGGGTSNFLIPNGTFFVVLLIFLIVLGVI
MSMEG_4940|M.smegmatis_MC2_155 -MGEFSAT----ILAASQAAEEGGGGSNFLIPNGTFFAVLIIFLIVLGVI
MMAR_4091|M.marinum_M -MDDVNSI----VLAAGQAAEEG-GTNNFLVPNGTFFFVLAIFLVVLAVI
MUL_3958|M.ulcerans_Agy99 -MDDVNSI----VLAAGQAAEEG-GTNNFLVPNGTFFFVLAIFLVVLAVI
Mb1338|M.bovis_AF2122/97 -MGEVSAI----VLAASQAAEEGGESSNFLIPNGTFFVVLAIFLVVLAVI
Rv1306|M.tuberculosis_H37Rv -MGEVSAI----VLAASQAAEEGGESSNFLIPNGTFFVVLAIFLVVLAVI
MLBr_01141|M.leprae_Br4923 -MFEVSAI----VFAVSQAAEEG-KSSNFLLPNGTFFFVLAIFLVVLGVI
MAV_1523|M.avium_104 MMGDASLS----VLASSQVVAEG--GNNFLVPNGTFFFVLAIFLIVLAVI
MAB_1449|M.abscessus_ATCC_1997 -MGELHSVASAVTAVAAEAAEEGGKQNNFLIPNGTFFVVLAIFLIVLAVI
TH_1562|M.thermoresistible__bu -----------------VAQEDGGTQN-FLLPNGTFFFTLLIFLIVLGVI
. :* . **:****** .* ***:**.**
Mflv_2314|M.gilvum_PYR-GCK AKWVVPPISKVLAEREAMLAKTAADNRKSAEQVAAARADYDKTLAEARGE
Mvan_4332|M.vanbaalenii_PYR-1 AKWVVPPVSKVLAEREAMLAKTAADNRKSAEQVAAAQADYDKTMADARGE
MSMEG_4940|M.smegmatis_MC2_155 SKWVVPPISKVLAEREAMLAKTAADNRKSAEQVAAAQADYEKEMAEARAQ
MMAR_4091|M.marinum_M GTFVVPPILKVLRERDAMVAKTLADNKKSAEQFAAAQADYEKAMAEARVQ
MUL_3958|M.ulcerans_Agy99 GTFVVPPILKVLRERDAMVAMTLADNKKSAEQFAAAQADYEKAMAEARVQ
Mb1338|M.bovis_AF2122/97 GTFVVPPILKVLRERDAMVAKTLADNKKSDEQFAAAQADYDEAMTEARVQ
Rv1306|M.tuberculosis_H37Rv GTFVVPPILKVLRERDAMVAKTLADNKKSDEQFAAAQADYDEAMTEARVQ
MLBr_01141|M.leprae_Br4923 GTFVVPPILKVLQERDAMVAKTDADSKMSAAQFAAAQADYEAAMKEARVQ
MAV_1523|M.avium_104 GTFVVPPVMKVLRERDAMVAKTAADNRKAAEQFEAAQADYEEAMTEARVQ
MAB_1449|M.abscessus_ATCC_1997 GTFVVPPIQKVLKAREDMVTKTAEDNRNAAEQFTAAEADYKDELAKARGA
TH_1562|M.thermoresistible__bu TKWVVPPINKVLREREAMVQKTAEDNRKSAELAAATEADTRRVLDAARRE
.:****: *** *: *: * *.: : *:.** : **
Mflv_2314|M.gilvum_PYR-GCK ASSIRDEARVAGRQVVDEKRATANGEVAETVKTADEKLTQQGSAAQSELQ
Mvan_4332|M.vanbaalenii_PYR-1 ASSIRDEARVAGRQVVDEKRAVASGEVAETVKSADQQLSQQGSAAQSELQ
MSMEG_4940|M.smegmatis_MC2_155 ASALRDEARAAGRSVVDEKRAQASGEVAQTLTQADQQLSAQGDQVRSGLE
MMAR_4091|M.marinum_M ASSYRDNARAEGRKVVEDARAHAEQEVASTLQQANEQLKRERDAVELDLR
MUL_3958|M.ulcerans_Agy99 ASSYRHNARAEGRKVVEDARAHAEQEVASTLQQANEQLKRERDAVELDLR
Mb1338|M.bovis_AF2122/97 ASSLRDNARADGRKVIEDARVRAEQQVASTLQTAHEQLKRERDAVELDLR
Rv1306|M.tuberculosis_H37Rv ASSLRDNARADGRKVIEDARVRAEQQVASTLQTAHEQLKRERDAVELDLR
MLBr_01141|M.leprae_Br4923 SSFLRDNARVDGRKSIEEARVRAEQHVVSTLQIAGEQIKRERDAVELDLR
MAV_1523|M.avium_104 ASSLRDNARAEGRKVVEDARAKAEQEVLSTLQLAARQLKRERDAVELDLR
MAB_1449|M.abscessus_ATCC_1997 ATAVRDEARAEGRGILEDMRQRANAEATAVTETAAAELARQGEVTAGELA
TH_1562|M.thermoresistible__bu ASATRDEARNQGRAVLEDMRSRASEEAAATLKTASEELSRQGEATRAELE
:: *.:** ** ::: * *. .. . * :: : . . *
Mflv_2314|M.gilvum_PYR-GCK SSVDALSATLASRILGVDVN--TRGSQ---------
Mvan_4332|M.vanbaalenii_PYR-1 SSVDGLSATLASRILGVDVN--SGGSR---------
MSMEG_4940|M.smegmatis_MC2_155 SSVDGLSAKLASRILGVDVN--SGGTQ---------
MMAR_4091|M.marinum_M ANVGAMSATLANRIVGVDVT--TPAAAG--------
MUL_3958|M.ulcerans_Agy99 ANVGAMSATLANRIVGVDVT--TPAAAG--------
Mb1338|M.bovis_AF2122/97 AHVGTMSATLASRILGVDLT--ASAATR--------
Rv1306|M.tuberculosis_H37Rv AHVGTMSATLASRILGVDLT--ASAATR--------
MLBr_01141|M.leprae_Br4923 AKAGAMSLILASRILGVDIT--ASVVTR--------
MAV_1523|M.avium_104 ANVASMSATLASRILGVDVA--PAAATTSATKTSGR
MAB_1449|M.abscessus_ATCC_1997 TNVDSLSRTLAERVLGVSLS--EPANAGRG------
TH_1562|M.thermoresistible__bu ASVEALSTKLAGRVLGIDVGGRTSATTGQGR-----
: . :* ** *::*:.: