For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MEQPRRTCHLVIGYRASLRMLAAGLVSQRGSFDRANAALAIDLARRHNPDELIAEFAELAVHPRGIGRVF PRRLLLGDHVIHELDIAFALDRTPTVGIDALVAVLNTQVRVPNPFVPAAQRARGLSLRAHDVDWTSGEGD WAVTGGAAQLASALAGRPWALDQLSGKGVDTLRTRMLLH
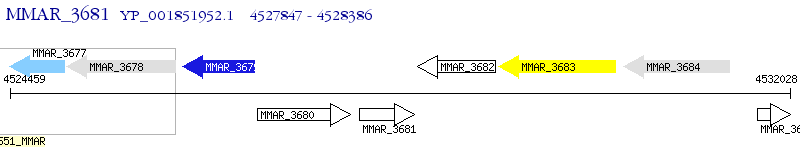
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3681 | - | - | 100% (179) | hypothetical protein MMAR_3681 |
| M. marinum M | MMAR_0893 | - | 2e-06 | 33.94% (165) | hypothetical protein MMAR_0893 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2062 | - | 3e-15 | 37.16% (148) | hypothetical protein Mb2062 |
| M. gilvum PYR-GCK | - | - | - | - | - |
| M. tuberculosis H37Rv | Rv2036 | - | 3e-15 | 37.84% (148) | hypothetical protein Rv2036 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4788c | - | 5e-18 | 39.47% (152) | hypothetical protein MAB_4788c |
| M. avium 104 | MAV_0227 | - | 4e-16 | 39.87% (158) | hypothetical protein MAV_0227 |
| M. smegmatis MC2 155 | MSMEG_6779 | - | 4e-05 | 31.61% (174) | hypothetical protein MSMEG_6779 |
| M. thermoresistible (build 8) | TH_1248 | - | 3e-15 | 36.24% (149) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_3624 | - | 3e-92 | 98.82% (169) | hypothetical protein MUL_3624 |
| M. vanbaalenii PYR-1 | Mvan_5391 | - | 2e-10 | 34.15% (164) | hypothetical protein Mvan_5391 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2062|M.bovis_AF2122/97 MIAADDDTEKSMMDMARAERAELAAFLTTLTLQQWETPSLCAGWSVKEVV
Rv2036|M.tuberculosis_H37Rv MIAADDDTEKSMMDMARAERAELAAFLTTLTLQQWETPSLCAGWSVKEVV
MAV_0227|M.avium_104 -------MTSELMAMARAERADLAEFLATLGPRDWEAPSLCSRWSVKDVV
TH_1248|M.thermoresistible__bu -----------MMDMAYAERTELAQFLATLTPEQWATPSLCARWTVKDVV
Mvan_5391|M.vanbaalenii_PYR-1 ---------------------------------------------MRDVA
MSMEG_6779|M.smegmatis_MC2_155 -MRLRTAERDRVFDAVAHERRGIADLIDGLTPEQLATPSLCGGWDVKTVA
MAB_4788c|M.abscessus_ATCC_199 ------MSRARVYAMVQQEKRDLAALLRELTPQEWESPSLCAGWRVRDVV
MMAR_3681|M.marinum_M -------------------------------------------MEQPRRT
MUL_3624|M.ulcerans_Agy99 --------------------------------------------------
Mb2062|M.bovis_AF2122/97 AHMISYEDLGVFGLLKRFAK--GRIVRANEVGVDEFAGLSPQE--LVDYV
Rv2036|M.tuberculosis_H37Rv AHMISYEDLGVFGLLKRFAK--GRIVRANEVGVDEFAGLSPQE--LADYV
MAV_0227|M.avium_104 AHVISYEELGAFGLLKRFAK--GWVVRANQVGVDEFADLTPQQ--LLEFL
TH_1248|M.thermoresistible__bu AHVISYEELGPFGLLKRFAK--GYVVRANQVGVDEFGSLSPEQ--LLDFL
Mvan_5391|M.vanbaalenii_PYR-1 AHTVGYLGQSVPGLIRNMIRDRGDVDRLNARMLPAVAALTPAE--LVELM
MSMEG_6779|M.smegmatis_MC2_155 AHLVSDFLDGFRGFLVSGIR-HGNMDKGIDALARRRAQAPAAE--IAETL
MAB_4788c|M.abscessus_ATCC_199 AHVL-YDGTPLLRYGSEVIRVRGSADRLNRLYLDRARGWSTEE--LLAAF
MMAR_3681|M.marinum_M CHLVIGYRASLRMLAAGLVSQRGSFDRANAALAIDLARRHNPDELIAEFA
MUL_3624|M.ulcerans_Agy99 ---MIGYRASLRMLAADLVSQRGSFDRANAALAIDLARRHNPDELIAEFA
: * : : :
Mb2062|M.bovis_AF2122/97 GRHLQPRGLTAGFGGMIALVDGMIHHQDIRRPLGQPRTIPAQRLDRVLRL
Rv2036|M.tuberculosis_H37Rv GRHLQPRGLTAGFGGMIALVDGMIHHQDIRRPLGQPRTIPAQRLDRVLRL
MAV_0227|M.avium_104 REHLEPRGLTAGFGGMIALVDGTIHHQDIRRALGRPRVVPPDRLQRVLSL
TH_1248|M.thermoresistible__bu NRHLWPRGLTAGFGGMIALVDGTVHHQDIRRPLGRPRTIAADRLLRVLRS
Mvan_5391|M.vanbaalenii_PYR-1 GRDSTPTGAAGLYGGRVALIECVIHQQDIRRPLGLDFDVPEDSLRVSLDY
MSMEG_6779|M.smegmatis_MC2_155 RTRAEFRLSPPVTGPLAGLTDVLVHGADIRIPLGLAHRPDPANVAWVLDF
MAB_4788c|M.abscessus_ATCC_199 EATIA-RSYSARIQPKLVLADLLIHQQDIRRPLNRLRTVPEGTLRMVLDN
MMAR_3681|M.marinum_M ELAVHPRGIGRVFPRRLLLGDHVIHELDIAFALDRTPTVGIDALVAVLNT
MUL_3624|M.ulcerans_Agy99 ELAVHPRGIGQVFPRRLLLGDHVIHELDIAFALDRTPTVGIDALVAVLNT
* : :* ** .*. : *
Mb2062|M.bovis_AF2122/97 MPKNPRLR--ARPRIKGLRLRATDLDWTIGTG-PEVTGPGEALLMAMAGR
Rv2036|M.tuberculosis_H37Rv MPKNPRLR--ARPRIKGLRLRATDLDWTIGTG-PEVTGPGEALLMAMAGR
MAV_0227|M.avium_104 VPGNPRLG--AGRRIRGLRLRATDIDWTHGDG-PQVSGPGEALLMAMSGR
TH_1248|M.thermoresistible__bu VPGNPRLG--AGRRIRGLRLHATDIDWTHGEG-PEVSGTGEALLMAMTGR
Mvan_5391|M.vanbaalenii_PYR-1 ARISPVIG--GTRRTRGLRLVATDMDWSAGTG-PEVCGTAEALLLAMTGR
MSMEG_6779|M.smegmatis_MC2_155 LAGRTQFGFFPQRRLRGLALHDGDTGRTWGAG-QEIRGPGVALMLAVCGR
MAB_4788c|M.abscessus_ATCC_199 P--DPFIN--PKRRLKGLRWTATDIQWTDGTG-PEVHGPGEAIVMAVGGR
MMAR_3681|M.marinum_M QVRVPNPFVPAAQRARGLSLRAHDVDWTSGEGDWAVTGGAAQLASALAGR
MUL_3624|M.ulcerans_Agy99 QVRVPNPFVPAAQRARGLSLRAHDVDWTSGEGDWAVTGGAAQLASALAGR
. * :** * : * * : * . : *: **
Mb2062|M.bovis_AF2122/97 P-AAVSDLSGPGKPTLAGRLG--
Rv2036|M.tuberculosis_H37Rv P-AAVSDLSGPGKPTLAGRLG--
MAV_0227|M.avium_104 P-AALADLDGPGRATLAARLGQ-
TH_1248|M.thermoresistible__bu A-AVVDELGGPGKPVLAGRLR--
Mvan_5391|M.vanbaalenii_PYR-1 ADAVRAELSGEGIPHLR------
MSMEG_6779|M.smegmatis_MC2_155 T-VALDRLTGPGVTVLRSRLT--
MAB_4788c|M.abscessus_ATCC_199 P-AALAQLDGPGVEILRTRLDGD
MMAR_3681|M.marinum_M P-WALDQLSGKGVDTLRTRMLLH
MUL_3624|M.ulcerans_Agy99 P-WALDQLSGKGVDTLRTRMLLH
. . * * * *