For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSVDAPNTAAEQGPEILTPDVFREFMSSYPSGVTIVTTYDQHGAPRGFTCSSMCSVTMEPPTLLLCVNTR STTLDAVQDCTRFAVNLLHSGGESAARVFSSVQEDRFSKVRWRASTIGGLPILYEDAHAIAECRLTATRN VGTHTVLFGEIDAIILDHQPLHYRADMNPLLYCRRQFTAFPTQQP
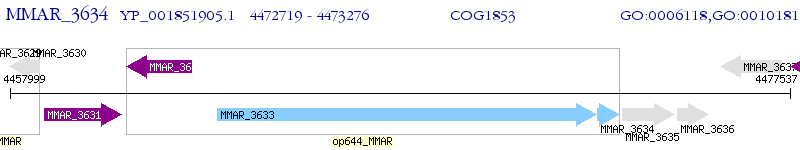
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. marinum M | MMAR_3634 | - | - | 100% (185) | oxidase |
| M. marinum M | MMAR_5062 | - | 7e-18 | 30.25% (162) | oxidase |
| M. marinum M | MMAR_2867 | - | 3e-11 | 31.45% (159) | oxidoreducatse |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3598c | - | 3e-18 | 29.17% (168) | oxidoreductase |
| M. gilvum PYR-GCK | Mflv_1466 | - | 2e-17 | 28.92% (166) | flavin reductase domain-containing protein |
| M. tuberculosis H37Rv | Rv3567c | - | 3e-18 | 29.17% (168) | oxidoreductase |
| M. leprae Br4923 | MLBr_02561 | - | 2e-08 | 21.74% (161) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0586 | - | 2e-17 | 30.30% (165) | oxidoreductase |
| M. avium 104 | MAV_0594 | - | 2e-17 | 29.63% (162) | putative flavin reductase |
| M. smegmatis MC2 155 | MSMEG_6035 | - | 2e-18 | 29.01% (162) | nitrilotriacetate monooxygenase component B |
| M. thermoresistible (build 8) | TH_1739 | - | 9e-19 | 29.07% (172) | POSSIBLE OXIDOREDUCTASE |
| M. ulcerans Agy99 | MUL_4138 | - | 3e-18 | 30.25% (162) | oxidase |
| M. vanbaalenii PYR-1 | Mvan_5305 | - | 2e-17 | 29.01% (162) | flavin reductase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1466|M.gilvum_PYR-GCK -----------MGSPPIDPRTFRNVLGQFCTGITVITTVHDD-APVGFAC
Mvan_5305|M.vanbaalenii_PYR-1 -----------MSKPPIDPRTFRNVLGQFCTGITVITTVHDG-SPVGFAC
MSMEG_6035|M.smegmatis_MC2_155 ----------MTATQPIDPRTFRNVLGQFCTGVTVITTVHEN-VPVGFAC
TH_1739|M.thermoresistible__bu -----------VTAEAIDQRTFRRVLGQFCTGVTIITTVHEG-NPVGFAC
Mb3598c|M.bovis_AF2122/97 -----------MSA-QIDPRTFRSVLGQFCTGITVITTVHDD-VPVGFAC
Rv3567c|M.tuberculosis_H37Rv -----------MSA-QIDPRTFRSVLGQFCTGITVITTVHDD-VPVGFAC
MAV_0594|M.avium_104 -----------MAA-PIDPRTFRQVLGQFCTGITIITTVHDE-VPVGFAC
MUL_4138|M.ulcerans_Agy99 -----------MAADPIDPRTFRHVLGQFCTGITIITTVHDD-VPSGFAC
MAB_0586|M.abscessus_ATCC_1997 -----------MSTPEIDPRTFRNVLGQFCTGITIITTTHED-VPVGFAC
MLBr_02561|M.leprae_Br4923 ----------MRAMNSLTPSLLRDAFGHFPSGVVAIAAEVEG-VREGLAA
MMAR_3634|M.marinum_M MSVDAPNTAAEQGPEILTPDVFREFMSSYPSGVTIVTTYDQHGAPRGFTC
: :* :. : :*:. ::: : *::.
Mflv_1466|M.gilvum_PYR-GCK QSFAALSLDPPLVLFCPTKASRSWAAIEASGTFCVNILHENQQHVSARFG
Mvan_5305|M.vanbaalenii_PYR-1 QSFAALSLDPPLVLFCPTKASRSWAAIEASGTFCVNILHENQKDVSARFG
MSMEG_6035|M.smegmatis_MC2_155 QSFAALSLDPPLVLFCPTKQSRAWKAIEATGRFCVNMLHENQQHVSAQFG
TH_1739|M.thermoresistible__bu QSFAALSLDPPLVLFCPTKVSRSWKAIEASGRFCVNILHEKQQHVSARFG
Mb3598c|M.bovis_AF2122/97 QSFAALSLEPPLVLFCPTKVSRSWQAIEASGRFCVNVLTEKQKDVSARFG
Rv3567c|M.tuberculosis_H37Rv QSFAALSLEPPLVLFCPTKVSRSWQAIEASGRFCVNVLTEKQKDVSARFG
MAV_0594|M.avium_104 QSFAALSLDPPLVLFCPTKVSRSWKAIEATGRFCVNVLTEKQKHVSARFG
MUL_4138|M.ulcerans_Agy99 QSFAALSLDPPLVLFCPTKVSRSWKAIEASGRFCVNVLTEKQKSVSARFG
MAB_0586|M.abscessus_ATCC_1997 QSFAALSLEPPLVLFCPTKQSRSWAAIAASGKFCVNVLAEDQREVSARFG
MLBr_02561|M.leprae_Br4923 STFVPVSLEPPLVSFCVQNVSTTWLKLKAVPMLGISVLGEVHDEVACTLA
MMAR_3634|M.marinum_M SSMCSVTMEPPTLLLCVNTRSTTLDAVQDCTRFAVNLLHSGGESAARVFS
.:: .::::** : :* . * : : : :.:* . .: :.
Mflv_1466|M.gilvum_PYR-GCK SREPDKFAGIDWAPSKLG--SPVIEGSLAHIDCTVHSVHDGGDHFVVFGA
Mvan_5305|M.vanbaalenii_PYR-1 SREPDKFAGIDWTPSKTG--SPIIDGTLAHIDCSVHSVHDGGDHFVVFGA
MSMEG_6035|M.smegmatis_MC2_155 SKAADKFAGIDWHPSKLG--SPVIDGTLAHIDCTVHSVHDGGDHFVVFGA
TH_1739|M.thermoresistible__bu SREPDKFAGIDWRPSDLG--SPIIDGSLAHIDCTVHDVHDGGDHFVVFGK
Mb3598c|M.bovis_AF2122/97 SKEPDKFAGIDWRPSELG--SPIIEGSLAYIDCTVASVHDGGDHFVVFGA
Rv3567c|M.tuberculosis_H37Rv SKEPDKFAGIDWRPSELG--SPIIEGSLAYIDCTVASVHDGGDHFVVFGA
MAV_0594|M.avium_104 SKEPDKFAGIDWHPSELG--SPIIDGALAHIDCTVASVHEGGDHFVVFGA
MUL_4138|M.ulcerans_Agy99 SKESDKFAGIDWRPSELG--SPIIDGSLAYIDCTVASVHDGGDHFVVFGA
MAB_0586|M.abscessus_ATCC_1997 SREPDKFAGINWRPSGLG--SPIINGSLAHIDCEVANVHDGGDHWVVFGS
MLBr_02561|M.leprae_Br4923 TKHGDRFAGLETVSNDTG--AVFIKGTSLWLESAIKQLIPAGDHTIVVLR
MMAR_3634|M.marinum_M SVQEDRFSKVRWRASTIGGLPILYEDAHAIAECRLTATRNVGTHTVLFGE
: *:*: : .. * . . ..: :. : * * ::.
Mflv_1466|M.gilvum_PYR-GCK VHSLSD---VPKKK--PRPLLFYHGQYTGIEPDKNTPADWRNDLEAFLTA
Mvan_5305|M.vanbaalenii_PYR-1 VHSLSD---VPKTK--PRPLLFYQGQYTGIEPDKNTPADWRNDLEAFLTA
MSMEG_6035|M.smegmatis_MC2_155 VHSLSE---VPKTK--PRPLLFYRGEYTGIEPDKNTPAQWRDDLEAFLTA
TH_1739|M.thermoresistible__bu VHGLSE---VPERK--PRPLLFYRGEYTGIEPEKNTPAQWRDDLEAFLTA
Mb3598c|M.bovis_AF2122/97 VESLSE---VPAVK--PRPLLFYRGDYTGIEPEKTTPAHWRDDLEAFLTT
Rv3567c|M.tuberculosis_H37Rv VESLSE---VPAVK--PRPLLFYRGDYTGIEPEKTTPAHWRDDLEAFLIT
MAV_0594|M.avium_104 VQSLSE---APKIK--PRPLLFYRGEYTGIEPDKTTPAQWRDDLEAFLTT
MUL_4138|M.ulcerans_Agy99 VHSLSE---VPKIK--PRPLLFYRGDYTGIEPDKTTPAQWRDDLEAFLTA
MAB_0586|M.abscessus_ATCC_1997 VKELSERAVIPSAESAARPLLFYRGQYTGIEPDKNTPADWRNDLEAFLTA
MLBr_02561|M.leprae_Br4923 VSQVTVD---PDVA----PIVFHRSMFRRLGF------------------
MMAR_3634|M.marinum_M IDAIILDHQPLHYRADMNPLLYCRRQFTAFPTQQP---------------
: : *::: : : :
Mflv_1466|M.gilvum_PYR-GCK TTDDTWL
Mvan_5305|M.vanbaalenii_PYR-1 TTDDTWL
MSMEG_6035|M.smegmatis_MC2_155 TTDDTWL
TH_1739|M.thermoresistible__bu TTADTWL
Mb3598c|M.bovis_AF2122/97 TTQDTWL
Rv3567c|M.tuberculosis_H37Rv TTQDTWL
MAV_0594|M.avium_104 TTQDTWL
MUL_4138|M.ulcerans_Agy99 TTQDTWL
MAB_0586|M.abscessus_ATCC_1997 TTEDTWL
MLBr_02561|M.leprae_Br4923 -------
MMAR_3634|M.marinum_M -------